High expression of long non-coding RNA NNT-AS1 facilitates progression of cholangiocarcinoma through promoting epithelial-mesenchymal transition
- PMID: 31632521
- PMCID: PMC6789241
High expression of long non-coding RNA NNT-AS1 facilitates progression of cholangiocarcinoma through promoting epithelial-mesenchymal transition
Abstract
Background: Cholangiocarcinoma (CCA) is a biliary malignancy, which is notoriously difficult to diagnose and associated with poor survival. Accumulating evidence indicates that long non-coding RNA Nicotinamide Nucleotide Transhydrogenase-antisense RNA1 (NNT-AS1) is overexpressed in several tumors and plays a crucial role in the development of neoplasm. However, the expression pattern and functional role of NNT-AS1 in CCA remain largely unknown.
Methods: NNT-AS1 expression was assessed by RT-qPCR and In Situ Hybridization (ISH) assay. The clinical relevance of NNT-AS1 was analyzed using a CCA tissue microarray with follow-up data. The function role of NNT-AS1 and its underlying molecular mechanisms were evaluated using both in vitro/in vivo experiments and bioinformatics analysis. Luciferase reporter assay, western blot and RT-qPCR were conducted to identify the miRNA/target gene involved in the regulation of CCA progression.
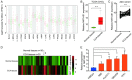
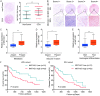
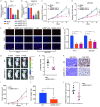
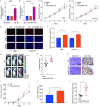
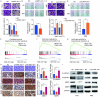
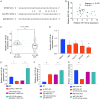
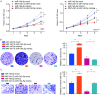
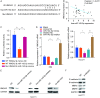
Results: LncRNA NNT-AS1 was found highly expressed in CCA. Upregulated NNT-AS1 expression was tightly associated with clinical malignancies and predicted poor prognosis of CCA patients. Functional studies showed that NNT-AS1 knockdown inhibited cell proliferation, migration and invasion of CCA cells in vitro. Conversely, NNT-AS1 overexpression showed the opposite biological effects. In a tumor xenograft model, we confirmed that NNT-AS1 knockdown could significantly inhibit the growth of CCA, while NNT-AS1 overexpression promoted CCA development. Mechanistically, we demonstrated that NNT-AS1 might function as a ceRNA in regulating HMGA2 (high mobility group AT-hook 2) through competitively binding to miR-142-5p in CCA. Moreover, we showed that NNT-AS1 regulated epithelial-mesenchymal transition in CCA.
Conclusion: In summary, these findings suggest the potential prognostic and therapeutic value of NNT-AS1/miR-142-5p/HMGA2 axis in CCA patients.
Keywords: NNT-AS1; cholangiocarcinoma; epithelial-mesenchymal transition; proliferation; tumor progression.
AJTR Copyright © 2019.
Conflict of interest statement
None.
Figures
Similar articles
-
Long non-coding RNA NNT-AS1 promotes cholangiocarcinoma cells proliferation and epithelial-to-mesenchymal transition through down-regulating miR-203.Aging (Albany NY). 2020 Feb 5;12(3):2333-2346. doi: 10.18632/aging.102747. Epub 2020 Feb 5. Aging (Albany NY). 2020. PMID: 32019904 Free PMC article.
-
Long non-coding RNA NNT-AS1 functions as an oncogenic gene through modulating miR-485/BCL9 in cholangiocarcinoma.Cancer Manag Res. 2019 Aug 15;11:7739-7749. doi: 10.2147/CMAR.S207801. eCollection 2019. Cancer Manag Res. 2019. PMID: 31616187 Free PMC article.
-
Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis.Oncotarget. 2017 Sep 28;8(51):88804-88814. doi: 10.18632/oncotarget.21321. eCollection 2017 Oct 24. Oncotarget. 2017. PMID: 29179477 Free PMC article.
-
The Role of Long Non-Coding RNA NNT-AS1 in Neoplastic Disease.Cancers (Basel). 2020 Oct 23;12(11):3086. doi: 10.3390/cancers12113086. Cancers (Basel). 2020. PMID: 33113895 Free PMC article. Review.
-
The Advancement of Long Non-Coding RNAs in Cholangiocarcinoma Development.J Cancer. 2019 May 26;10(11):2407-2414. doi: 10.7150/jca.32411. eCollection 2019. J Cancer. 2019. PMID: 31258745 Free PMC article. Review.
Cited by
-
Long Non-Coding RNAs as Molecular Biomarkers in Cholangiocarcinoma.Front Cell Dev Biol. 2022 Apr 27;10:890605. doi: 10.3389/fcell.2022.890605. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35573683 Free PMC article. Review.
-
Emerging roles of long noncoding RNAs in cholangiocarcinoma: Advances and challenges.Cancer Commun (Lond). 2020 Dec;40(12):655-680. doi: 10.1002/cac2.12109. Epub 2020 Nov 3. Cancer Commun (Lond). 2020. PMID: 33142045 Free PMC article. Review.
-
Role of long non-coding RNAs in cholangiocarcinoma: A systematic review and meta-analysis.Cancer Rep (Hoboken). 2024 Mar;7(3):e2029. doi: 10.1002/cnr2.2029. Cancer Rep (Hoboken). 2024. PMID: 38517409 Free PMC article.
-
Extracellular vesicle-derived miRNA-mediated cell-cell communication inference for single-cell transcriptomic data with miRTalk.Genome Biol. 2025 Apr 14;26(1):95. doi: 10.1186/s13059-025-03566-x. Genome Biol. 2025. PMID: 40229908 Free PMC article.
-
Upregulated expression of LncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1 is correlated with unfavorable clinical outcomes in cancers.BMC Cancer. 2020 Sep 14;20(1):879. doi: 10.1186/s12885-020-07348-5. BMC Cancer. 2020. PMID: 32928135 Free PMC article.
References
-
- Alberts SR, Gores GJ, Kim GP, Roberts LR, Kendrick ML, Rosen CB, Chari ST, Martenson JA. Treatment options for hepatobiliary and pancreatic cancer. Mayo Clin Proc. 2007;82:628–637. - PubMed
LinkOut - more resources
Full Text Sources
