Variable levels of drift in tunicate cardiopharyngeal gene regulatory elements
- PMID: 31632631
- PMCID: PMC6790052
- DOI: 10.1186/s13227-019-0137-2
Variable levels of drift in tunicate cardiopharyngeal gene regulatory elements
Abstract
Background: Mutations in gene regulatory networks often lead to genetic divergence without impacting gene expression or developmental patterning. The rules governing this process of developmental systems drift, including the variable impact of selective constraints on different nodes in a gene regulatory network, remain poorly delineated.
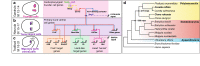
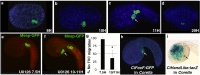
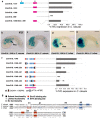
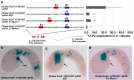
Results: Here we examine developmental systems drift within the cardiopharyngeal gene regulatory networks of two tunicate species, Corella inflata and Ciona robusta. Cross-species analysis of regulatory elements suggests that trans-regulatory architecture is largely conserved between these highly divergent species. In contrast, cis-regulatory elements within this network exhibit distinct levels of conservation. In particular, while most of the regulatory elements we analyzed showed extensive rearrangements of functional binding sites, the enhancer for the cardiopharyngeal transcription factor FoxF is remarkably well-conserved. Even minor alterations in spacing between binding sites lead to loss of FoxF enhancer function, suggesting that bound trans-factors form position-dependent complexes.
Conclusions: Our findings reveal heterogeneous levels of divergence across cardiopharyngeal cis-regulatory elements. These distinct levels of divergence presumably reflect constraints that are not clearly associated with gene function or position within the regulatory network. Thus, levels of cis-regulatory divergence or drift appear to be governed by distinct structural constraints that will be difficult to predict based on network architecture.
Keywords: Developmental systems drift; Gene regulatory networks; Heart development; Selective constraints; Tunicates.
© The Author(s) 2019.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

