MARTINI-Based Protein-DNA Coarse-Grained HADDOCKing
- PMID: 31632986
- PMCID: PMC6779769
- DOI: 10.3389/fmolb.2019.00102
MARTINI-Based Protein-DNA Coarse-Grained HADDOCKing
Abstract
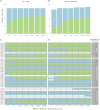
Modeling biomolecular assemblies is an important field in computational structural biology. The inherent complexity of their energy landscape and the computational cost associated with modeling large and complex assemblies are major drawbacks for integrative modeling approaches. The so-called coarse-graining approaches, which reduce the degrees of freedom of the system by grouping several atoms into larger "pseudo-atoms," have been shown to alleviate some of those limitations, facilitating the identification of the global energy minima assumed to correspond to the native state of the complex, while making the calculations more efficient. Here, we describe and assess the implementation of the MARTINI force field for DNA into HADDOCK, our integrative modeling platform. We combine it with our previous implementation for protein-protein coarse-grained docking, enabling coarse-grained modeling of protein-nucleic acid complexes. The system is modeled using MARTINI topologies and interaction parameters during the rigid body docking and semi-flexible refinement stages of HADDOCK, and the resulting models are then converted back to atomistic resolution by an atom-to-bead distance restraints-guided protocol. We first demonstrate the performance of this protocol using 44 complexes from the protein-DNA docking benchmark, which shows an overall ~6-fold speed increase and maintains similar accuracy as compared to standard atomistic calculations. As a proof of concept, we then model the interaction between the PRC1 and the nucleosome (a former CAPRI target in round 31), using the same information available at the time the target was offered, and compare all-atom and coarse-grained models.
Keywords: biomolecular complexes; coarse-graining; docking; force field; nucleic acids.
Copyright © 2019 Honorato, Roel-Touris and Bonvin.
Figures
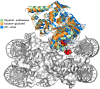
References
LinkOut - more resources
Full Text Sources
Research Materials

