ROMOP: a light-weight R package for interfacing with OMOP-formatted electronic health record data
- PMID: 31633087
- PMCID: PMC6800657
- DOI: 10.1093/jamiaopen/ooy059
ROMOP: a light-weight R package for interfacing with OMOP-formatted electronic health record data
Abstract
Objectives: Electronic health record (EHR) data are increasingly used for biomedical discoveries. The nature of the data, however, requires expertise in both data science and EHR structure. The Observational Medical Out-comes Partnership (OMOP) common data model (CDM) standardizes the language and structure of EHR data to promote interoperability of EHR data for research. While the OMOP CDM is valuable and more attuned to research purposes, it still requires extensive domain knowledge to utilize effectively, potentially limiting more widespread adoption of EHR data for research and quality improvement.
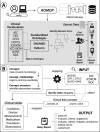
Materials and methods: We have created ROMOP: an R package for direct interfacing with EHR data in the OMOP CDM format.
Results: ROMOP streamlines typical EHR-related data processes. Its functions include exploration of data types, extraction and summarization of patient clinical and demographic data, and patient searches using any CDM vocabulary concept.
Conclusion: ROMOP is freely available under the Massachusetts Institute of Technology (MIT) license and can be obtained from GitHub (http://github.com/BenGlicksberg/ROMOP). We detail instructions for setup and use in the Supplementary Materials. Additionally, we provide a public sandbox server containing synthesized clinical data for users to explore OMOP data and ROMOP (http://romop.ucsf.edu).
Conflict of interest statement
Conflict of interest statement. None declared.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
