High-throughput micropatterning platform reveals Nodal-dependent bisection of peri-gastrulation-associated versus preneurulation-associated fate patterning
- PMID: 31634368
- PMCID: PMC6822778
- DOI: 10.1371/journal.pbio.3000081
High-throughput micropatterning platform reveals Nodal-dependent bisection of peri-gastrulation-associated versus preneurulation-associated fate patterning
Abstract
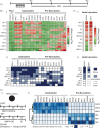
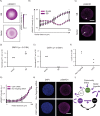
In vitro models of postimplantation human development are valuable to the fields of regenerative medicine and developmental biology. Here, we report characterization of a robust in vitro platform that enabled high-content screening of multiple human pluripotent stem cell (hPSC) lines for their ability to undergo peri-gastrulation-like fate patterning upon bone morphogenetic protein 4 (BMP4) treatment of geometrically confined colonies and observed significant heterogeneity in their differentiation propensities along a gastrulation associable and neuralization associable axis. This cell line-associated heterogeneity was found to be attributable to endogenous Nodal expression, with up-regulation of Nodal correlated with expression of a gastrulation-associated gene profile, and Nodal down-regulation correlated with a preneurulation-associated gene profile expression. We harness this knowledge to establish a platform of preneurulation-like fate patterning in geometrically confined hPSC colonies in which fates arise because of a BMPs signalling gradient conveying positional information. Our work identifies a Nodal signalling-dependent switch in peri-gastrulation versus preneurulation-associated fate patterning in hPSC cells, provides a technology to robustly assay hPSC differentiation outcomes, and suggests conserved mechanisms of organized fate specification in differentiating epiblast and ectodermal tissues.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Hertig AT, Rock J, Adams EC. A description of 34 human ova within the first 17 days of development. Am J Anat. 1956;98(3):435–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

