The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis
- PMID: 31634900
- PMCID: PMC6883167
- DOI: 10.1038/s41586-019-1705-2
The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis
Abstract
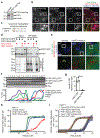
Ferroptosis is a form of regulated cell death that is caused by the iron-dependent peroxidation of lipids1,2. The glutathione-dependent lipid hydroperoxidase glutathione peroxidase 4 (GPX4) prevents ferroptosis by converting lipid hydroperoxides into non-toxic lipid alcohols3,4. Ferroptosis has previously been implicated in the cell death that underlies several degenerative conditions2, and induction of ferroptosis by the inhibition of GPX4 has emerged as a therapeutic strategy to trigger cancer cell death5. However, sensitivity to GPX4 inhibitors varies greatly across cancer cell lines6, which suggests that additional factors govern resistance to ferroptosis. Here, using a synthetic lethal CRISPR-Cas9 screen, we identify ferroptosis suppressor protein 1 (FSP1) (previously known as apoptosis-inducing factor mitochondrial 2 (AIFM2)) as a potent ferroptosis-resistance factor. Our data indicate that myristoylation recruits FSP1 to the plasma membrane where it functions as an oxidoreductase that reduces coenzyme Q10 (CoQ) (also known as ubiquinone-10), which acts as a lipophilic radical-trapping antioxidant that halts the propagation of lipid peroxides. We further find that FSP1 expression positively correlates with ferroptosis resistance across hundreds of cancer cell lines, and that FSP1 mediates resistance to ferroptosis in lung cancer cells in culture and in mouse tumour xenografts. Thus, our data identify FSP1 as a key component of a non-mitochondrial CoQ antioxidant system that acts in parallel to the canonical glutathione-based GPX4 pathway. These findings define a ferroptosis suppression pathway and indicate that pharmacological inhibition of FSP1 may provide an effective strategy to sensitize cancer cells to ferroptosis-inducing chemotherapeutic agents.
Figures






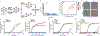

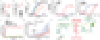
Comment in
-
A powerful cell-protection system prevents cell death by ferroptosis.Nature. 2019 Nov;575(7784):597-598. doi: 10.1038/d41586-019-03145-8. Nature. 2019. PMID: 31768036 Free PMC article.
-
A new checkpoint against ferroptosis.Cell Res. 2020 Jan;30(1):3-4. doi: 10.1038/s41422-019-0258-0. Cell Res. 2020. PMID: 31772274 Free PMC article. No abstract available.
-
The Antioxidant Role of Non-mitochondrial CoQ10: Mystery Solved!Cell Metab. 2020 Jan 7;31(1):13-15. doi: 10.1016/j.cmet.2019.12.007. Cell Metab. 2020. PMID: 31951565
References
-
- Ingold I et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 172, 409–422.e21 (2018). - PubMed
-
- Dixon SJ & Stockwell BR The hallmarks of ferroptosis. Annu. Rev. Cancer Biol 3, 35–54 (2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

