Viral Innate Immune Evasion and the Pathogenesis of Emerging RNA Virus Infections
- PMID: 31635238
- PMCID: PMC6832425
- DOI: 10.3390/v11100961
Viral Innate Immune Evasion and the Pathogenesis of Emerging RNA Virus Infections
Abstract
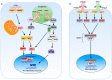
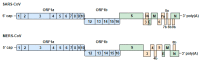
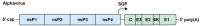
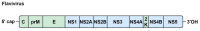
Positive-sense single-stranded RNA (+ssRNA) viruses comprise many (re-)emerging human pathogens that pose a public health problem. Our innate immune system and, in particular, the interferon response form the important first line of defence against these viruses. Given their genetic flexibility, these viruses have therefore developed multiple strategies to evade the innate immune response in order to optimize their replication capacity. Already many molecular mechanisms of innate immune evasion by +ssRNA viruses have been identified. However, research addressing the effect of host innate immune evasion on the pathology caused by viral infections is less prevalent in the literature, though very relevant and interesting. Since interferons have been implicated in inflammatory diseases and immunopathology in addition to their protective role in infection, antagonizing the immune response may have an ambiguous effect on the clinical outcome of the viral disease. Therefore, this review discusses what is currently known about the role of interferons and host immune evasion in the pathogenesis of emerging coronaviruses, alphaviruses and flaviviruses.
Keywords: innate immune evasion; positive-sense single-stranded RNA viruses; type I and III interferons; viral pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- WHO Middle East respiratory syndrome coronavirus (MERS-CoV) [(accessed on 16 September 2019)]; Available online: https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory...
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

