GenBank is a reliable resource for 21st century biodiversity research
- PMID: 31636175
- PMCID: PMC6842603
- DOI: 10.1073/pnas.1911714116
GenBank is a reliable resource for 21st century biodiversity research
Abstract
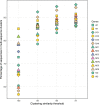
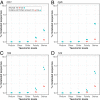
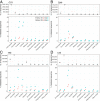
Traditional methods of characterizing biodiversity are increasingly being supplemented and replaced by approaches based on DNA sequencing alone. These approaches commonly involve extraction and high-throughput sequencing of bulk samples from biologically complex communities or samples of environmental DNA (eDNA). In such cases, vouchers for individual organisms are rarely obtained, often unidentifiable, or unavailable. Thus, identifying these sequences typically relies on comparisons with sequences from genetic databases, particularly GenBank. While concerns have been raised about biases and inaccuracies in laboratory and analytical methods, comparatively little attention has been paid to the taxonomic reliability of GenBank itself. Here we analyze the metazoan mitochondrial sequences of GenBank using a combination of distance-based clustering and phylogenetic analysis. Because of their comparatively rapid evolutionary rates and consequent high taxonomic resolution, mitochondrial sequences represent an invaluable resource for the detection of the many small and often undescribed organisms that represent the bulk of animal diversity. We show that metazoan identifications in GenBank are surprisingly accurate, even at low taxonomic levels (likely <1% error rate at the genus level). This stands in contrast to previously voiced concerns based on limited analyses of particular groups and the fact that individual researchers currently submit annotated sequences to GenBank without significant external taxonomic validation. Our encouraging results suggest that the rapid uptake of DNA-based approaches is supported by a bioinformatic infrastructure capable of assessing both the losses to biodiversity caused by global change and the effectiveness of conservation efforts aimed at slowing or reversing these losses.
Keywords: environmental DNA; metabarcoding; taxonomic assignments.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

Comment in
-
Reply to Locatelli et al.: Evaluating species-level accuracy of GenBank metazoan sequences will require experts' effort in each group.Proc Natl Acad Sci U S A. 2020 Dec 22;117(51):32213-32214. doi: 10.1073/pnas.2019903117. Epub 2020 Nov 24. Proc Natl Acad Sci U S A. 2020. PMID: 33234564 Free PMC article. No abstract available.
-
GenBank's reliability is uncertain for biodiversity researchers seeking species-level assignment for eDNA.Proc Natl Acad Sci U S A. 2020 Dec 22;117(51):32211-32212. doi: 10.1073/pnas.2007421117. Epub 2020 Nov 24. Proc Natl Acad Sci U S A. 2020. PMID: 33234565 Free PMC article. No abstract available.
References
-
- Bohmann K., et al. , Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367 (2014). - PubMed
-
- Creer S., et al. , The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol. Evol. 7, 1008–1018 (2016).
-
- Adamowicz S. J., et al. , Trends in DNA barcoding and metabarcoding. Genome 62, v–viii (2019). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

