Unified rational protein engineering with sequence-based deep representation learning
- PMID: 31636460
- PMCID: PMC7067682
- DOI: 10.1038/s41592-019-0598-1
Unified rational protein engineering with sequence-based deep representation learning
Abstract
Rational protein engineering requires a holistic understanding of protein function. Here, we apply deep learning to unlabeled amino-acid sequences to distill the fundamental features of a protein into a statistical representation that is semantically rich and structurally, evolutionarily and biophysically grounded. We show that the simplest models built on top of this unified representation (UniRep) are broadly applicable and generalize to unseen regions of sequence space. Our data-driven approach predicts the stability of natural and de novo designed proteins, and the quantitative function of molecularly diverse mutants, competitively with the state-of-the-art methods. UniRep further enables two orders of magnitude efficiency improvement in a protein engineering task. UniRep is a versatile summary of fundamental protein features that can be applied across protein engineering informatics.
Figures

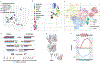

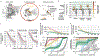
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

