Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways
- PMID: 31637532
- PMCID: PMC6803609
- DOI: 10.1186/s12284-019-0334-6
Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways
Abstract
Background: The NAC (NAM, AFAT, and CUC) transcription factors play critical roles in rice (Oryza sativa) development and stress regulation. Overexpressing a microRNA (miR164b)-resistant OsNAC2 mutant gene, which generates transcripts that cannot be targeted by miR164b, improves rice plant architecture and yield; however, the performance of these mOsNAC2-overexpressing lines, named ZUOErN3 and ZUOErN4, under abiotic stress conditions such as drought have not yet been fully characterized.
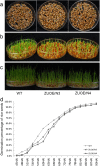
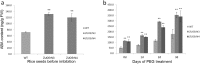
Results: In this study, we showed that the germination of ZUOErN3 and ZUOErN4 seeds was delayed in comparison with the wild-type (WT) seeds, although the final germination rates of all lines were over 95%. The quantification of the endogenous ABA levels revealed that the germinating mOsNAC2-overexpressing seeds had elevated ABA levels, which resulted in their slower germination. The mOsNAC2-overexpressing plants were significantly more drought tolerance than the WT plants, with the survival rate increasing from 11.2% in the WT to nearly 70% in ZUOErN3 and ZUOErN4 plants after a drought treatment. Salt (NaCl) tolerance was also increased in the ZUOErN3 and ZUOErN4 plants due to significantly increased ABA levels. A reverse transcription quantitative PCR (RT-qPCR) analysis showed a significant increase in the expression of the ABA biosynthesis genes OsNCED1 and OsNCED3 in the mOsNAC2-overexpressing lines, and the expression levels of the stress-responsive genes OsP5CS1, OsLEA3, and OsRab16 were significantly increased in these plants. Moreover, OsNAC2 directly interacted with the promoters of OsLEA3 and OsNCED3 in yeast one-hybrid assays.
Conclusions: Taken together, our results show that OsNAC2 plays a positive regulatory role in drought and salt tolerance in rice through ABA-mediated pathways.
Keywords: ABA; Drought tolerance; OsNAC2; Rice (Oryza sativa); Salt tolerance; microRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Do TH, Michel J, Geert A, Christian H, Tran TT, Le VS, Nancy HR. Proline accumulation and Δ1-pyrroline-5-carboxylate synthetase gene properties in three rice cultivars differing in salinity and drought tolerance. Plant Sci. 2003;165(5):1059–1068. doi: 10.1016/S0168-9452(03)00301-7. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

