Salt Dependence of A-Form RNA Duplexes: Structures and Implications
- PMID: 31638810
- PMCID: PMC7068736
- DOI: 10.1021/acs.jpcb.9b07502
Salt Dependence of A-Form RNA Duplexes: Structures and Implications
Abstract
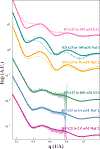
The biological functions of RNA range from gene regulation through catalysis and depend critically on its structure and flexibility. Conformational variations of flexible, non-base-paired components, including RNA hinges, bulges, or single-stranded tails, are well documented. Recent work has also identified variations in the structure of ubiquitous, base-paired duplexes found in almost all functional RNAs. Duplexes anchor the structures of folded RNAs, and their surface features are recognized by partner molecules. To date, no consistent picture has been obtained that describes the range of conformations assumed by RNA duplexes. Here, we apply wide angle, solution X-ray scattering (WAXS) to quantify these variations, by sampling length scales characteristic of helical geometries under different solution conditions. To identify the radius, helical rise, twist, and length of dsRNA helices, we exploit molecular dynamics generated structures, explicit solvent models, and ensemble optimization methods. Our results quantify the substantial and salt-dependent deviations of double-stranded (ds) RNA duplexes from the assumed canonical A-form conformation. Recent experiments underscore the need to properly describe the structures of RNA duplexes when interpreting the salt dependence of RNA conformations.
Figures


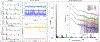

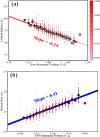
References
-
- Dock-Bregeon AC, Chevrier B, Podjarny A, Moras D, deBear JS, Gough GR, Gilham PT and Johnson JE High resolution structure of the RNA duplex [U(U-A)6A]2. Nature. 335(6188): 375–8 (1988). - PubMed
-
- Holbrook SR, Cheong C, Tinoco I and Kim SH Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 353(6344): 579–81 (1991). - PubMed
-
- Tolbert BS, Miyazaki Y, Barton S, Kinde B, Starck P, Singh R, Bax A, Case DA and Summers MF Major groove width variations in RNA structures determined by NMR and impact of13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement. J. Biomol. NMR 47(3):205–19 (2010). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

