A framework for TRIM21-mediated protein depletion in early mouse embryos: recapitulation of Tead4 null phenotype over three days
- PMID: 31638890
- PMCID: PMC6805607
- DOI: 10.1186/s12864-019-6106-2
A framework for TRIM21-mediated protein depletion in early mouse embryos: recapitulation of Tead4 null phenotype over three days
Abstract
Background: While DNA and RNA methods are routine to disrupt the expression of specific genes, complete understanding of developmental processes requires also protein methods, because: oocytes and early embryos accumulate proteins and these are not directly affected by DNA and RNA methods. When proteins in the oocyte encounter a specific antibody and the TRIpartite Motiv-containing 21 (TRIM21) ubiquitin-protein ligase, they can be committed to degradation in the proteasome, producing a transient functional knock-out that reveals the role of the protein. However, there are doubts about whether this targeted proteolysis could be successfully used to study mammalian development, because duration of the transient effect is unknown, and also because amounts of reagents delivered must be adequate in relation to the amount of target protein, which is unknown, too.
Results: We show that the mouse egg contains up to 1E-02 picomoles/protein, as estimated by mass spectrometry using the intensity-based absolute quantification (iBAQ) algorithm. However, the egg can only accommodate ≈1E-04 picomoles of antibody or TRIM21 without incurring toxic effects. Within this framework, we demonstrate that TRIM21-mediated protein depletion efficiently disrupts the embryonic process of trophectoderm formation, which critically depends on the TEA domain family member 4 (Tead4) gene. TEAD4 depletion starting at the 1-cell stage lasts for 3 days prior to a return of gene and protein expression to baseline. This time period is long enough to result in a phenotype entirely consistent with that of the published null mutation and RNA interference studies: significant underexpression of trophectodermal genes Cdx2 and Gata3 and strongly impaired ability of embryos to cavitate and implant in the uterus. Omics data are available via ProteomeXchange (PXD012613) and GEO (GSE124844).
Conclusions: TRIM21-mediated protein depletion can be an effective means to disrupt gene function in mouse development, provided the target gene is chosen carefully and the method is tuned accurately. The knowledge gathered in this study provides the basic know-how (prerequisites, requirements, limitations) to expedite the protein depletion of other genes besides Tead4.
Keywords: Mouse; Oocyte; Preimplantation embryo; Proteome; TEAD4; TRIM21; Trophectoderm.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
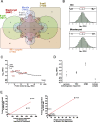

 ), anti-GFP antibody (
), anti-GFP antibody ( ) or anti-TEAD4 antibody (
) or anti-TEAD4 antibody ( ), compared to the background fluorescence of non-manipulated cells (
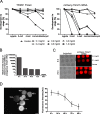
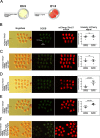
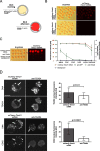
), compared to the background fluorescence of non-manipulated cells ( ). N = 3 zygotes or embryos per stage per treatment. Note the secondary right axis used in plot to better discern the background fluorescence values. d. Representative immunofluorescent signals (largest cross section, nucleus fluorescence) of TEAD4 and CDX2 in TRIM21-only and TEAD4-depleted embryos at day E3.5 (n = 7 TEAD4-depleted and n = 8 TRIM21-only embryos for TEAD4 immunofluorescence; n = 11 TEAD-depleted and n = 8 TRIM21-only embryos for CDX2 immunofluorescence). DNA stained with YO-PRO-1. Arrows point at peripheral nuclei that are TEAD4- or CDX2-positive in controls but negative in TEAD4-depleted embryos. Size bar, 50 μm. OGDB, Oregon Green dextran beads. Error bars = standard deviations. Statistical significance tested with Student’s t test. AU, arbitrary units of fluorescence intensity
). N = 3 zygotes or embryos per stage per treatment. Note the secondary right axis used in plot to better discern the background fluorescence values. d. Representative immunofluorescent signals (largest cross section, nucleus fluorescence) of TEAD4 and CDX2 in TRIM21-only and TEAD4-depleted embryos at day E3.5 (n = 7 TEAD4-depleted and n = 8 TRIM21-only embryos for TEAD4 immunofluorescence; n = 11 TEAD-depleted and n = 8 TRIM21-only embryos for CDX2 immunofluorescence). DNA stained with YO-PRO-1. Arrows point at peripheral nuclei that are TEAD4- or CDX2-positive in controls but negative in TEAD4-depleted embryos. Size bar, 50 μm. OGDB, Oregon Green dextran beads. Error bars = standard deviations. Statistical significance tested with Student’s t test. AU, arbitrary units of fluorescence intensity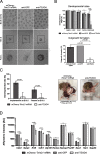
Similar articles
-
TEAD4 regulates trophectoderm differentiation upstream of CDX2 in a GATA3-independent manner in the human preimplantation embryo.Hum Reprod. 2022 Jul 30;37(8):1760-1773. doi: 10.1093/humrep/deac138. Hum Reprod. 2022. PMID: 35700449
-
Effects of downregulating TEAD4 transcripts by RNA interference on early development of bovine embryos.J Reprod Dev. 2017 Apr 21;63(2):135-142. doi: 10.1262/jrd.2016-130. Epub 2016 Dec 11. J Reprod Dev. 2017. PMID: 27941302 Free PMC article.
-
Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development.Development. 2007 Nov;134(21):3827-36. doi: 10.1242/dev.010223. Epub 2007 Oct 3. Development. 2007. PMID: 17913785
-
Maternal factors regulating preimplantation development in mice.Curr Top Dev Biol. 2020;140:317-340. doi: 10.1016/bs.ctdb.2019.10.006. Epub 2019 Nov 19. Curr Top Dev Biol. 2020. PMID: 32591079 Free PMC article. Review.
-
Mechanisms of trophectoderm fate specification in preimplantation mouse development.Dev Growth Differ. 2010 Apr;52(3):263-73. doi: 10.1111/j.1440-169X.2009.01158.x. Epub 2010 Jan 20. Dev Growth Differ. 2010. PMID: 20100249 Review.
Cited by
-
Comparative maternal protein profiling of mouse biparental and uniparental embryos.Gigascience. 2022 Sep 3;11:giac084. doi: 10.1093/gigascience/giac084. Gigascience. 2022. PMID: 36056732 Free PMC article.
-
Protein-targeting reverse genetic approaches: the future of oocyte and preimplantation embryo research.Mol Hum Reprod. 2025 Apr 3;31(2):gaaf008. doi: 10.1093/molehr/gaaf008. Mol Hum Reprod. 2025. PMID: 40100642 Review.
-
Elevated RIF1 participates in the epigenetic abnormalities of zygotes by regulating histone modifications on MuERV-L in obese mice.Mol Med. 2022 Feb 5;28(1):17. doi: 10.1186/s10020-022-00446-z. Mol Med. 2022. PMID: 35123389 Free PMC article.
-
Initiation of a conserved trophectoderm program in human, cow and mouse embryos.Nature. 2020 Nov;587(7834):443-447. doi: 10.1038/s41586-020-2759-x. Epub 2020 Sep 23. Nature. 2020. PMID: 32968278 Free PMC article.
-
Targeted protein degradation in mammalian cells: A promising avenue toward future.Comput Struct Biotechnol J. 2022 Sep 28;20:5477-5489. doi: 10.1016/j.csbj.2022.09.038. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36249565 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

