The interplay between miR156/SPL13 and DFR/WD40-1 regulate drought tolerance in alfalfa
- PMID: 31638916
- PMCID: PMC6802326
- DOI: 10.1186/s12870-019-2059-5
The interplay between miR156/SPL13 and DFR/WD40-1 regulate drought tolerance in alfalfa
Abstract
Background: Developing Medicago sativa L. (alfalfa) cultivars tolerant to drought is critical for the crop's sustainable production. miR156 regulates various plant biological functions by silencing SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors.
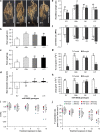
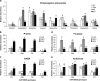
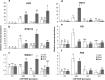
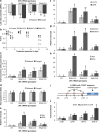
Results: To understand the mechanism of miR156-modulated drought stress tolerance in alfalfa we used genotypes with altered expression levels of miR156, miR156-regulated SPL13, and DIHYDROFLAVONOL-4-REDUCTASE (DFR) regulating WD40-1. Previously we reported the involvement of miR156 in drought tolerance, but the mechanism and downstream genes involved in this process were not fully studied. Here we illustrate the interplay between miR156/SPL13 and WD40-1/DFR to regulate drought stress by coordinating gene expression with metabolite and physiological strategies. Low to moderate levels of miR156 overexpression suppressed SPL13 and increased WD40-1 to fine-tune DFR expression for enhanced anthocyanin biosynthesis. This, in combination with other accumulated stress mitigating metabolites and physiological responses, improved drought tolerance. We also demonstrated that SPL13 binds in vivo to the DFR promoter to regulate its expression.
Conclusions: Taken together, our results reveal that moderate relative miR156 transcript levels are sufficient to enhance drought resilience in alfalfa by silencing SPL13 and increasing WD40-1 expression, whereas higher miR156 overexpression results in drought susceptibility.
Keywords: Alfalfa; Drought; SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE13; WD40–1; miR156; microRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Olesen JE, Trnka M, Kersebaum KC, Skjelvåg AO, Seguin B, Peltonen-Sainio P, et al. Impacts and adaptation of European crop production systems to climate change. Eur J Agron. 2011;34(2):96–112. doi: 10.1016/j.eja.2010.11.003. - DOI
-
- Mba C, Guimaraes EP, Ghosh K. Re-orienting crop improvement for the changing climatic conditions of the 21st century. Agric Food Secur. 2012;1(1):1–17. doi: 10.1186/2048-7010-1-7. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

