The R882H substitution in the human de novo DNA methyltransferase DNMT3A disrupts allosteric regulation by the tumor supressor p53
- PMID: 31640986
- PMCID: PMC6885622
- DOI: 10.1074/jbc.RA119.010827
The R882H substitution in the human de novo DNA methyltransferase DNMT3A disrupts allosteric regulation by the tumor supressor p53
Abstract
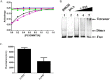
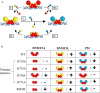
A myriad of protein partners modulate the activity of the human DNA methyltransferase 3A (DNMT3A), whose interactions with these other proteins are frequently altered during oncogenesis. We show here that the tumor suppressor p53 decreases DNMT3A activity by forming a heterotetramer complex with DNMT3A. Mutational and modeling experiments suggested that p53 interacts with the same region in DNMT3A as does the structurally characterized DNMT3L. We observed that the p53-mediated repression of DNMT3A activity is blocked by amino acid substitutions within this interface, but surprisingly, also by a distal DNMT3A residue, R882H. DNMT3A R882H occurs frequently in various cancers, including acute myeloid leukemia, and our results suggest that the effects of R882H and other DNMT3A mutations may go beyond changes in DNMT3A methylation activity. To further understand the dynamics of how protein-protein interactions modulate DNMT3A activity, we determined that p53 has a greater affinity for DNMT3A than for DNMT3L and that p53 readily displaces DNMT3L from the DNMT3A:DNMT3L heterotetramer. Interestingly, this occurred even when the preformed DNMT3A:DNMT3L complex was actively methylating DNA. The frequently identified p53 substitutions (R248W and R273H), whereas able to regulate DNMT3A function when forming the DNMT3A:p53 heterotetramer, no longer displaced DNMT3L from the DNMT3A:DNMT3L heterotetramer. The results of our work highlight the complex interplay between DNMT3A, p53, and DNMT3L and how these interactions are further modulated by clinically derived mutations in each of the interacting partners.
Keywords: DNA demethylation; DNMT3A; allosteric regulation; epigenetics; nucleic acid enzymology; p53; protein-protein interaction.
© 2019 Sandoval and Reich.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
References
-
- Vertino P. M., Sekowski J. A., Coll J. M., Applegreen N., Han S., Hickey R. J., and Malkas L. H. (2002) DNMT1 is a component of a multiprotein DNA replication complex. Cell Cycle 1, 416–423 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

