Human VH1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis
- PMID: 31641091
- PMCID: PMC6805997
- DOI: 10.1128/mBio.02473-19
Human VH1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis
Abstract
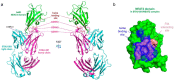
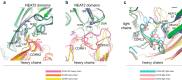
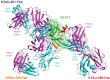
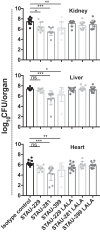
Staphylococcus aureus is an important human pathogen that infects nearly every human tissue. Like most organisms, the acquisition of nutrient iron is necessary for its survival. One route by which it obtains this metal is through the iron-regulated surface determinant (Isd) system that scavenges iron from the hemoglobin of the host. We show that the heavy chain variable region IGHV1-69 gene commonly encodes human monoclonal antibodies (mAbs) targeting IsdB-NEAT2. Remarkably, these antibodies bind to multiple antigenic sites. One class of IGHV1-69-encoded mAbs blocks S. aureus heme acquisition by binding to the heme-binding site of NEAT2, while two additional classes reduce the bacterial burden in vivo by an alternative Fc receptor-mediated mechanism. We further identified clonal lineages of IGHV1-69-encoded mAbs using donor samples, showing that each lineage diversifies during infection by somatic hypermutation. These studies reveal that IGHV1-69-encoded antibodies contribute to a protective immune response, furthering our understanding of the correlates of protection against S. aureus infection.IMPORTANCE The human pathogen Staphylococcus aureus causes a wide range of infections, including skin abscesses and sepsis. There is currently no licensed vaccine to prevent S. aureus infection, and its treatment has become increasingly difficult due to antibiotic resistance. One potential way to inhibit S. aureus pathogenesis is to prevent iron acquisition. The iron-regulated surface determinant (Isd) system has evolved in S. aureus to acquire hemoglobin from the human host as a source of heme-iron. In this study, we investigated the molecular and structural basis for antibody-mediated correlates against a member of the Isd system, IsdB. The association of immunoglobulin heavy chain variable region IGHV1-69 gene-encoded human monoclonal antibodies with the response against S. aureus IsdB is described using structural and functional studies to define the importance of this antibody class. We also determine that somatic hypermutation in the development of these antibodies hinders rather than fine-tunes the immune response to IsdB.
Keywords: Staphylococcus aureus; X-ray crystallography; adaptive immunity; antibody functions; antibody repertoire; computer modeling; humoral immunity; monoclonal antibodies; proteomics.
Copyright © 2019 Bennett et al.
Figures



References
-
- Dillen CA, Pinsker BL, Marusina AI, Merleev AA, Farber ON, Liu H, Archer NK, Lee DB, Wang Y, Ortines RV, Lee SK, Marchitto MC, Cai SS, Ashbaugh AG, May LS, Holland SM, Freeman AF, Miller LG, Yeaman MR, Simon SI, Milner JD, Maverakis E, Miller LS. 2018. Clonally expanded gammadelta T cells protect against Staphylococcus aureus skin reinfection. J Clin Invest 128:1026–1042. doi: 10.1172/JCI96481. - DOI - PMC - PubMed
-
- Diep BA, Phung Q, Date S, Arnott D, Bakalarski C, Xu M, Nakamura G, Swem DL, Alexander MK, Le HN, Mai TT, Tan M-W, Brown EJ, Nishiyama M. 2014. Identifying potential therapeutic targets of methicillin-resistant Staphylococcus aureus through in vivo proteomic analysis. J Infect Dis 209:1533–1541. doi: 10.1093/infdis/jit662. - DOI - PMC - PubMed
-
- Bennett MR, Bombardi RG, Kose N, Parrish EH, Nagel MB, Petit RA, Read TD, Schey KL, Thomsen IP, Skaar EP, Crowe JE Jr.. 2019. Human mAbs to Staphylococcus aureus IsdA provide protection through both heme-blocking and Fc-mediated mechanisms. J Infect Dis 219:1264–1273. doi: 10.1093/infdis/jiy635. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
