Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in Zucker diabetic fatty rats
- PMID: 31641523
- PMCID: PMC6777410
- DOI: 10.1136/bmjdrc-2019-000717
Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in Zucker diabetic fatty rats
Abstract
Objective: Recent studies have demonstrated that gut microbiota was closely related to metabolic disorders such as type 2 diabetes. Oral antidiabetic medications including metformin, acarbose and sitagliptin lowered blood glucose levels via acting on the gastrointestinal tract. The aim of the study was to observe the comparisons among those medications on gut microbiota composition.
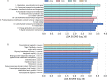
Research design and methods: Zucker diabetic fatty rats (n=32) were randomly divided into four groups, and had respectively gastric administration of normal saline (control), metformin (215.15 mg/kg/day), acarbose (32.27 mg/kg/day), or sitagliptin (10.76 mg/kg/day) for 4 weeks. Blood glucose levels were measured during an intragastric starch tolerance test after the treatments. 16S rRNA gene sequencing was used to access the microbiota in the fecal samples.
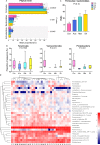
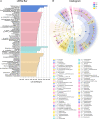
Results: Metformin, acarbose, and sitagliptin monotherapy effectively decreased fasting and postprandial blood glucose levels (p<0.001). Acarbose group displayed specific cluster and enterotype mainly composed by Ruminococcus 2 while Lactobacillus was the dominant bacterium in the enterotype of the other three groups. The relative abundance of genera Ruminococcus 2 and Bifidobacterium was dramatically higher in acarbose group. Metformin and sitagliptin increased the relative abundance of genus Lactobacillus. Metagenomic prediction showed that the functional profiles of carbohydrate metabolism were enriched in acarbose group.
Conclusions: Metformin, acarbose and sitagliptin exerted different effects on the composition of gut microbiota and selectively increased the beneficial bacteria. Supplementation with specific probiotics may further improve the hypoglycemic effects of the antidiabetic drugs.
Keywords: Metformin; acarbose; gut microbiota; sitagliptin; type 2 diabetes.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
