IDDomainSpotter: Compositional bias reveals domains in long disordered protein regions-Insights from transcription factors
- PMID: 31642121
- PMCID: PMC6933863
- DOI: 10.1002/pro.3754
IDDomainSpotter: Compositional bias reveals domains in long disordered protein regions-Insights from transcription factors
Abstract
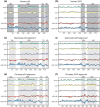
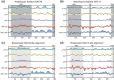
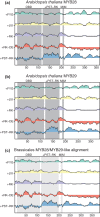
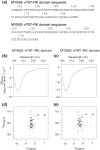
Protein domains constitute regions of distinct structural properties and molecular functions that are retained when removed from the rest of the protein. However, due to the lack of tertiary structure, the identification of domains has been largely neglected for long (>50 residues) intrinsically disordered regions. Here we present a sequence-based approach to assess and visualize domain organization in long intrinsically disordered regions based on compositional sequence biases. An online tool to find putative intrinsically disordered domains (IDDomainSpotter) in any protein sequence or sequence alignment using any particular sequence trait is available at http://www.bio.ku.dk/sbinlab/IDDomainSpotter. Using this tool, we have identified a putative domain enriched in hydrophilic and disorder-promoting residues (Pro, Ser, and Thr) and depleted in positive charges (Arg and Lys) bordering the folded DNA-binding domains of several transcription factors (p53, GCR, NAC46, MYB28, and MYB29). This domain, from two different MYB transcription factors, was characterized biophysically to determine its properties. Our analyses show the domain to be extended, dynamic and highly disordered. It connects the DNA-binding domain to other disordered domains and is present and conserved in several transcription factors from different families and domains of life. This example illustrates the potential of IDDomainSpotter to predict, from sequence alone, putative domains of functional interest in otherwise uncharacterized disordered proteins.
Keywords: DNA-binding domain; IDDomainSpotter; IDPs; NMR; compositional bias; domain; low-complexity regions; p53; plant MYB protein; transactivation domain; transcription factor.
© 2019 The Protein Society.
Figures
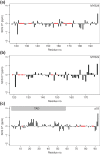
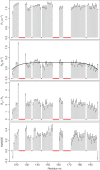
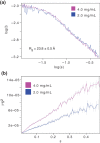

References
-
- Borcherds W, Theillet F‐X, Katzer A, et al. Disorder and residual helicity alter p53‐Mdm2 binding affinity and signaling in cells. Nat Chem Biol. 2014;10:1000–1002. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

