A computational method for design of connected catalytic networks in proteins
- PMID: 31642127
- PMCID: PMC6863703
- DOI: 10.1002/pro.3757
A computational method for design of connected catalytic networks in proteins
Abstract
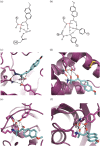
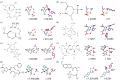
Computational design of new active sites has generally proceeded by geometrically defining interactions between the reaction transition state(s) and surrounding side-chain functional groups which maximize transition-state stabilization, and then searching for sites in protein scaffolds where the specified side-chain-transition-state interactions can be realized. A limitation of this approach is that the interactions between the side chains themselves are not constrained. An extensive connected hydrogen bond network involving the catalytic residues was observed in a designed retroaldolase following directed evolution. Such connected networks could increase catalytic activity by preorganizing active site residues in catalytically competent orientations, and enabling concerted interactions between side chains during catalysis, for example, proton shuffling. We developed a method for designing active sites in which the catalytic side chains, in addition to making interactions with the transition state, are also involved in extensive hydrogen bond networks. Because of the added constraint of hydrogen-bond connectivity between the catalytic side chains, to find solutions, a wider range of interactions between these side chains and the transition state must be considered. Our new method starts from a ChemDraw-like two-dimensional representation of the transition state with hydrogen-bond donors, acceptors, and covalent interaction sites indicated, and all placements of side-chain functional groups that make the indicated interactions with the transition state, and are fully connected in a single hydrogen-bond network are systematically enumerated. The RosettaMatch method can then be used to identify realizations of these fully-connected active sites in protein scaffolds. The method generates many fully-connected active site solutions for a set of model reactions that are promising starting points for the design of fully-preorganized enzyme catalysts.
Keywords: biocatalysis; computational modeling; enzyme design; enzyme mechanism; protein design.
© 2019 The Authors. Protein Science published by Wiley Periodicals, Inc. on behalf of The Protein Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Röthlisberger D, Khersonsky O, Wollacott AM, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

