The Genome Sequence of the Eastern Woodchuck (Marmota monax) - A Preclinical Animal Model for Chronic Hepatitis B
- PMID: 31645421
- PMCID: PMC6893209
- DOI: 10.1534/g3.119.400413
The Genome Sequence of the Eastern Woodchuck (Marmota monax) - A Preclinical Animal Model for Chronic Hepatitis B
Abstract
The Eastern woodchuck (Marmota monax) has been extensively used in research of chronic hepatitis B and liver cancer because its infection with the woodchuck hepatitis virus closely resembles a human hepatitis B virus infection. Development of novel immunotherapeutic approaches requires genetic information on immune pathway genes in this animal model. The woodchuck genome was assembled with a combination of high-coverage whole-genome shotgun sequencing of Illumina paired-end, mate-pair libraries and fosmid pool sequencing. The result is a 2.63 Gigabase (Gb) assembly with a contig N50 of 74.5 kilobases (kb), scaffold N50 of 892 kb, and genome completeness of 99.2%. RNA sequencing (RNA-seq) from seven different tissues aided in the annotation of 30,873 protein-coding genes, which in turn encode 41,826 unique protein products. More than 90% of the genes have been functionally annotated, with 82% of them containing open reading frames. This genome sequence and its annotation will enable further research in chronic hepatitis B and hepatocellular carcinoma and contribute to the understanding of immunological responses in the woodchuck.
Keywords: Chronic Hepatitis B; Eastern Woodchuck; Genome Assembly; Hepatocellular Carcinoma; Immune Response; Marmota monax; Whole Genome Sequencing.
Copyright © 2019 Alioto et al.
Figures
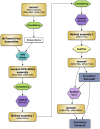
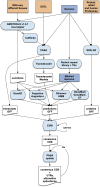
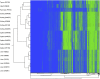
Similar articles
-
The genome of the American groundhog, Marmota monax.F1000Res. 2020 Sep 16;9:1137. doi: 10.12688/f1000research.25970.1. eCollection 2020. F1000Res. 2020. PMID: 33274050 Free PMC article.
-
Transcriptome Analysis and Comparison of Marmota monax and Marmota himalayana.PLoS One. 2016 Nov 2;11(11):e0165875. doi: 10.1371/journal.pone.0165875. eCollection 2016. PLoS One. 2016. PMID: 27806133 Free PMC article.
-
Characterization of interleukin 8 in woodchucks with chronic hepatitis B and hepatocellular carcinoma.Genes Immun. 2009 Jan;10(1):27-36. doi: 10.1038/gene.2008.86. Epub 2008 Oct 30. Genes Immun. 2009. PMID: 18971938
-
Immunomodulation as an option for the treatment of chronic hepatitis B virus infection: preclinical studies in the woodchuck model.Expert Opin Investig Drugs. 2007 Jun;16(6):787-801. doi: 10.1517/13543784.16.6.787. Expert Opin Investig Drugs. 2007. PMID: 17501692 Review.
-
Hepatocellular carcinoma in the woodchuck model of hepatitis B virus infection.Gastroenterology. 2004 Nov;127(5 Suppl 1):S283-93. doi: 10.1053/j.gastro.2004.09.043. Gastroenterology. 2004. PMID: 15508096 Review.
Cited by
-
Application of the woodchuck animal model for the treatment of hepatitis B virus-induced liver cancer.World J Gastrointest Oncol. 2021 Jun 15;13(6):509-535. doi: 10.4251/wjgo.v13.i6.509. World J Gastrointest Oncol. 2021. PMID: 34163570 Free PMC article. Review.
-
Imaging, Pathology, and Immune Correlates in the Woodchuck Hepatic Tumor Model.J Hepatocell Carcinoma. 2021 Mar 9;8:71-83. doi: 10.2147/JHC.S287800. eCollection 2021. J Hepatocell Carcinoma. 2021. PMID: 33728278 Free PMC article.
-
The genome of the American groundhog, Marmota monax.F1000Res. 2020 Sep 16;9:1137. doi: 10.12688/f1000research.25970.1. eCollection 2020. F1000Res. 2020. PMID: 33274050 Free PMC article.
-
A draft genome assembly for the eastern fox squirrel, Sciurus niger.G3 (Bethesda). 2021 Dec 8;11(12):jkab315. doi: 10.1093/g3journal/jkab315. G3 (Bethesda). 2021. PMID: 34550334 Free PMC article.
-
Forthcoming Developments in Models to Study the Hepatitis B Virus Replication Cycle, Pathogenesis, and Pharmacological Advancements.ACS Omega. 2023 Apr 14;8(16):14273-14289. doi: 10.1021/acsomega.2c07154. eCollection 2023 Apr 25. ACS Omega. 2023. PMID: 37125123 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
