Mitochondrial stress causes neuronal dysfunction via an ATF4-dependent increase in L-2-hydroxyglutarate
- PMID: 31645461
- PMCID: PMC6891100
- DOI: 10.1083/jcb.201904148
Mitochondrial stress causes neuronal dysfunction via an ATF4-dependent increase in L-2-hydroxyglutarate
Abstract
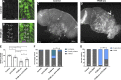
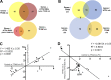
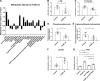
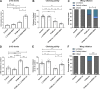
Mitochondrial stress contributes to a range of neurological diseases. Mitonuclear signaling pathways triggered by mitochondrial stress remodel cellular physiology and metabolism. How these signaling mechanisms contribute to neuronal dysfunction and disease is poorly understood. We find that mitochondrial stress in neurons activates the transcription factor ATF4 as part of the endoplasmic reticulum unfolded protein response (UPR) in Drosophila We show that ATF4 activation reprograms nuclear gene expression and contributes to neuronal dysfunction. Mitochondrial stress causes an ATF4-dependent increase in the level of the metabolite L-2-hydroxyglutarate (L-2-HG) in the Drosophila brain. Reducing L-2-HG levels directly, by overexpressing L-2-HG dehydrogenase, improves neurological function. Modulation of L-2-HG levels by mitochondrial stress signaling therefore regulates neuronal function.
© 2019 Hunt et al.
Figures
References
-
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Grüning B.A., et al. . 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46(W1):W537–W544. 10.1093/nar/gky379 - DOI - PMC - PubMed
-
- Benjamini Y., Krieger A.M., and Yekutieli D.. 2006. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 93:491–507. 10.1093/biomet/93.3.491 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

