Bacterial biodiversity drives the evolution of CRISPR-based phage resistance
- PMID: 31645729
- PMCID: PMC6837874
- DOI: 10.1038/s41586-019-1662-9
Bacterial biodiversity drives the evolution of CRISPR-based phage resistance
Abstract
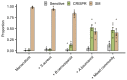
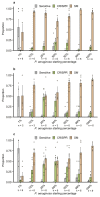
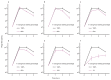
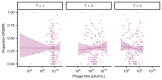
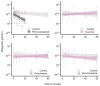
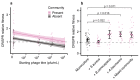
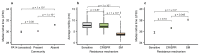
About half of all bacteria carry genes for CRISPR-Cas adaptive immune systems1, which provide immunological memory by inserting short DNA sequences from phage and other parasitic DNA elements into CRISPR loci on the host genome2. Whereas CRISPR loci evolve rapidly in natural environments3,4, bacterial species typically evolve phage resistance by the mutation or loss of phage receptors under laboratory conditions5,6. Here we report how this discrepancy may in part be explained by differences in the biotic complexity of in vitro and natural environments7,8. Specifically, by using the opportunistic pathogen Pseudomonas aeruginosa and its phage DMS3vir, we show that coexistence with other human pathogens amplifies the fitness trade-offs associated with the mutation of phage receptors, and therefore tips the balance in favour of the evolution of CRISPR-based resistance. We also demonstrate that this has important knock-on effects for the virulence of P. aeruginosa, which became attenuated only if the bacteria evolved surface-based resistance. Our data reveal that the biotic complexity of microbial communities in natural environments is an important driver of the evolution of CRISPR-Cas adaptive immunity, with key implications for bacterial fitness and virulence.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. - PubMed
-
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
-
- Westra ER, et al. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr Biol. 2015;25:1043–1049. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

