MHC-II neoantigens shape tumour immunity and response to immunotherapy
- PMID: 31645760
- PMCID: PMC6858572
- DOI: 10.1038/s41586-019-1671-8
MHC-II neoantigens shape tumour immunity and response to immunotherapy
Abstract
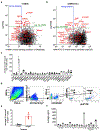
The ability of the immune system to eliminate and shape the immunogenicity of tumours defines the process of cancer immunoediting1. Immunotherapies such as those that target immune checkpoint molecules can be used to augment immune-mediated elimination of tumours and have resulted in durable responses in patients with cancer that did not respond to previous treatments. However, only a subset of patients benefit from immunotherapy and more knowledge about what is required for successful treatment is needed2-4. Although the role of tumour neoantigen-specific CD8+ T cells in tumour rejection is well established5-9, the roles of other subsets of T cells have received less attention. Here we show that spontaneous and immunotherapy-induced anti-tumour responses require the activity of both tumour-antigen-specific CD8+ and CD4+ T cells, even in tumours that do not express major histocompatibility complex (MHC) class II molecules. In addition, the expression of MHC class II-restricted antigens by tumour cells is required at the site of successful rejection, indicating that activation of CD4+ T cells must also occur in the tumour microenvironment. These findings suggest that MHC class II-restricted neoantigens have a key function in the anti-tumour response that is nonoverlapping with that of MHC class I-restricted neoantigens and therefore needs to be considered when identifying patients who will most benefit from immunotherapy.
Figures


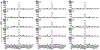







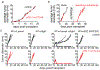



Comment in
-
Teamwork by different T-cell types boosts tumour destruction by immunotherapy.Nature. 2019 Oct;574(7780):639-640. doi: 10.1038/d41586-019-03106-1. Nature. 2019. PMID: 31659314 No abstract available.
-
Joint effort needed.Nat Rev Immunol. 2019 Dec;19(12):717. doi: 10.1038/s41577-019-0242-4. Nat Rev Immunol. 2019. PMID: 31666729 No abstract available.
References
-
- Schreiber RD, Old LJ & Smyth MJ Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570 (2011). - PubMed
METHODS ONLY REFERENCES
-
- Mamitsuka H Predicting peptides that bind to MHC molecules using supervised learning of hidden Markov models. Proteins 33, 460–474 (1998). - PubMed
-
- Welch LR Hidden Markov Models and the Baum-Welch Algorithm. IEEE Inf. Theory Soc. Newsl 53, 10–13 (2003).
-
- Kowalewski DJ & Stevanović S Biochemical large-scale identification of MHC class I ligands. Methods Mol. Biol. Clifton NJ 960, 145–157 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

