Evolution of the new head by gradual acquisition of neural crest regulatory circuits
- PMID: 31645763
- PMCID: PMC6858584
- DOI: 10.1038/s41586-019-1691-4
Evolution of the new head by gradual acquisition of neural crest regulatory circuits
Abstract
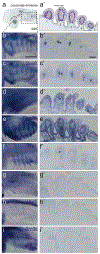
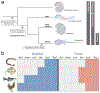
The neural crest, an embryonic stem-cell population, is a vertebrate innovation that has been proposed to be a key component of the 'new head', which imbued vertebrates with predatory behaviour1,2. Here, to investigate how the evolution of neural crest cells affected the vertebrate body plan, we examined the molecular circuits that control neural crest development along the anteroposterior axis of a jawless vertebrate, the sea lamprey. Gene expression analysis showed that the cranial subpopulation of the neural crest of the lamprey lacks most components of a transcriptional circuit that is specific to the cranial neural crest in amniotes and confers the ability to form craniofacial cartilage onto non-cranial neural crest subpopulations3. Consistent with this, hierarchical clustering analysis revealed that the transcriptional profile of the lamprey cranial neural crest is more similar to the trunk neural crest of amniotes. Notably, analysis of the cranial neural crest in little skate and zebrafish embryos demonstrated that the transcriptional circuit that is specific to the cranial neural crest emerged via the gradual addition of network components to the neural crest of gnathostomes, which subsequently became restricted to the cephalic region. Our results indicate that the ancestral neural crest at the base of the vertebrate lineage possessed a trunk-like identity. We propose that the emergence of the cranial neural crest, by progressive assembly of an axial-specific regulatory circuit, allowed the elaboration of the new head during vertebrate evolution.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



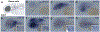




References
-
- Gans C & Northcutt RG Neural crest and the origin of vertebrates: a new head. Science 220, 268–273 (1983). - PubMed
-
- Glenn Northcutt R The new head hypothesis revisited. J. Exp. Zool. B Mol. Dev. Evol 304B, 274–297 (2005). - PubMed
-
- Le Douarin NM Development Of The Peripheral Nervous System From The Neural Crest. Annu. Rev. Cell Dev. Biol 4, 375–404 (1988). - PubMed
-
- Le Douarin N The Neural Crest. (Cambridge University Press, 1982).
Methods References:
-
- Nikitina N, Bronner-Fraser M & Sauka-Spengler T Culturing lamprey embryos. Cold Spring Harb Protoc 2009, pdb.prot5122–pdb.prot5122 (2009). - PubMed
-
- O’Sullivan O, Suhre K, Abergel C, Higgins DG & Notredame C 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol 340, 385–395 (2004). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

