Mechanisms by which PE21, an extract from the white willow Salix alba, delays chronological aging in budding yeast
- PMID: 31645900
- PMCID: PMC6791382
- DOI: 10.18632/oncotarget.27209
Mechanisms by which PE21, an extract from the white willow Salix alba, delays chronological aging in budding yeast
Abstract
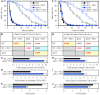
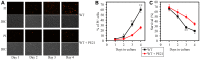
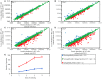
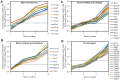
We have recently found that PE21, an extract from the white willow Salix alba, slows chronological aging and prolongs longevity of the yeast Saccharomyces cerevisiae more efficiently than any of the previously known pharmacological interventions. Here, we investigated mechanisms through which PE21 delays yeast chronological aging and extends yeast longevity. We show that PE21 causes a remodeling of lipid metabolism in chronologically aging yeast, thereby instigating changes in the concentrations of several lipid classes. We demonstrate that such changes in the cellular lipidome initiate three mechanisms of aging delay and longevity extension. The first mechanism through which PE21 slows aging and prolongs longevity consists in its ability to decrease the intracellular concentration of free fatty acids. This postpones an age-related onset of liponecrotic cell death promoted by excessive concentrations of free fatty acids. The second mechanism of aging delay and longevity extension by PE21 consists in its ability to decrease the concentrations of triacylglycerols and to increase the concentrations of glycerophospholipids within the endoplasmic reticulum membrane. This activates the unfolded protein response system in the endoplasmic reticulum, which then decelerates an age-related decline in protein and lipid homeostasis and slows down an aging-associated deterioration of cell resistance to stress. The third mechanisms underlying aging delay and longevity extension by PE21 consists in its ability to change lipid concentrations in the mitochondrial membranes. This alters certain catabolic and anabolic processes in mitochondria, thus amending the pattern of aging-associated changes in several key aspects of mitochondrial functionality.
Keywords: cellular aging; geroprotectors; lipid metabolism; mitochondria; necrotic cell death.
Copyright: © 2019 Medkour et al.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures
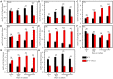
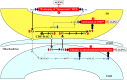

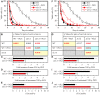
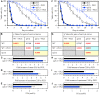






Similar articles
-
Caloric restriction delays yeast chronological aging by remodeling carbohydrate and lipid metabolism, altering peroxisomal and mitochondrial functionalities, and postponing the onsets of apoptotic and liponecrotic modes of regulated cell death.Oncotarget. 2018 Mar 5;9(22):16163-16184. doi: 10.18632/oncotarget.24604. eCollection 2018 Mar 23. Oncotarget. 2018. PMID: 29662634 Free PMC article.
-
Mechanisms through which lithocholic acid delays yeast chronological aging under caloric restriction conditions.Oncotarget. 2018 Oct 9;9(79):34945-34971. doi: 10.18632/oncotarget.26188. eCollection 2018 Oct 9. Oncotarget. 2018. PMID: 30405886 Free PMC article.
-
Lithocholic bile acid accumulated in yeast mitochondria orchestrates a development of an anti-aging cellular pattern by causing age-related changes in cellular proteome.Cell Cycle. 2015;14(11):1643-56. doi: 10.1080/15384101.2015.1026493. Cell Cycle. 2015. PMID: 25839782 Free PMC article.
-
Some Metabolites Act as Second Messengers in Yeast Chronological Aging.Int J Mol Sci. 2018 Mar 15;19(3):860. doi: 10.3390/ijms19030860. Int J Mol Sci. 2018. PMID: 29543708 Free PMC article. Review.
-
Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae.Microb Cell. 2014 May 27;1(6):163-178. doi: 10.15698/mic2014.06.152. Microb Cell. 2014. PMID: 28357241 Free PMC article. Review.
Cited by
-
Discovery of fifteen new geroprotective plant extracts and identification of cellular processes they affect to prolong the chronological lifespan of budding yeast.Oncotarget. 2020 Jun 9;11(23):2182-2203. doi: 10.18632/oncotarget.27615. eCollection 2020 Jun 9. Oncotarget. 2020. PMID: 32577164 Free PMC article.
-
Yeast Chronological Lifespan: Longevity Regulatory Genes and Mechanisms.Cells. 2022 May 23;11(10):1714. doi: 10.3390/cells11101714. Cells. 2022. PMID: 35626750 Free PMC article. Review.
-
Exploring the anti-aging potential of natural products and plant extracts in budding yeast Saccharomyces cerevisiae: A review.F1000Res. 2024 Dec 17;12:1265. doi: 10.12688/f1000research.141669.2. eCollection 2023. F1000Res. 2024. PMID: 39822944 Free PMC article. Review.
References
-
- Weissman J, Guthrie C, Fink GR. Guide to Yeast Genetics: Functional Genomics, Proteomics, and Other Systems Analyses Burlington: Academic Press; 2010. Available from: https://www.elsevier.com/books/guide-to-yeast-genetics-functional-genomi....
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

