C9orf72 expansion within astrocytes reduces metabolic flexibility in amyotrophic lateral sclerosis
- PMID: 31647549
- PMCID: PMC6906594
- DOI: 10.1093/brain/awz302
C9orf72 expansion within astrocytes reduces metabolic flexibility in amyotrophic lateral sclerosis
Abstract
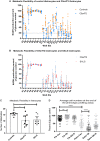
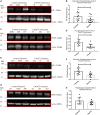
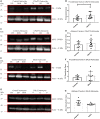
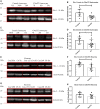
It is important to understand how the disease process affects the metabolic pathways in amyotrophic lateral sclerosis and whether these pathways can be manipulated to ameliorate disease progression. To analyse the basis of the metabolic defect in amyotrophic lateral sclerosis we used a phenotypic metabolic profiling approach. Using fibroblasts and reprogrammed induced astrocytes from C9orf72 and sporadic amyotrophic lateral sclerosis cases we measured the production rate of reduced nicotinamide adenine dinucleotides (NADH) from 91 potential energy substrates simultaneously. Our screening approach identified that C9orf72 and sporadic amyotrophic lateral sclerosis induced astrocytes have distinct metabolic profiles compared to controls and displayed a loss of metabolic flexibility that was not observed in fibroblast models. This loss of metabolic flexibility, involving defects in adenosine, fructose and glycogen metabolism, as well as disruptions in the membrane transport of mitochondrial specific energy substrates, contributed to increased starvation induced toxicity in C9orf72 induced astrocytes. A reduction in glycogen metabolism was attributed to loss of glycogen phosphorylase and phosphoglucomutase at the protein level in both C9orf72 induced astrocytes and induced neurons. In addition, we found alterations in the levels of fructose metabolism enzymes and a reduction in the methylglyoxal removal enzyme GLO1 in both C9orf72 and sporadic models of disease. Our data show that metabolic flexibility is important in the CNS in times of bioenergetic stress.
Keywords: C9orf72; ALS; astrocytes; metabolism; methylglyoxal.
© The Author(s) (2019). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures


References
-
- Allen SP, Duffy LM, Shaw PJ, Grierson AJ. Altered age-related changes in bioenergetic properties and mitochondrial morphology in fibroblasts from sporadic amyotrophic lateral sclerosis patients. Neurobiol Aging 2015; 36: 2893–903. - PubMed
-
- Allen S, Heath PR, Kirby J, Wharton SB, Cookson MR, Menzies FM, et al. Analysis of the cytosolic proteome in a cell culture model of familial amyotrophic lateral sclerosis reveals alterations to the proteasome, antioxidant defenses, and nitric oxide synthetic pathways. J Biol Chem 2003; 278: 6371–83. - PubMed
-
- Allen SP, Rajan S, Duffy L, Mortiboys H, Higginbottom A, Grierson AJ, et al. Superoxide dismutase 1 mutation in a cellular model of amyotrophic lateral sclerosis shifts energy generation from oxidative phosphorylation to glycolysis. Neurobiol Aging 2014; 35: 1499–509. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

