Transethnic meta-analysis of metabolic syndrome in a multiethnic study
- PMID: 31647587
- PMCID: PMC6986323
- DOI: 10.1002/gepi.22267
Transethnic meta-analysis of metabolic syndrome in a multiethnic study
Abstract
Genome-wide association studies (GWAS) have been used to establish thousands of genetic associations across numerous phenotypes. To improve the power of GWAS and generalize associations across ethnic groups, transethnic meta-analysis methods are used to combine the results of several GWAS from diverse ancestries. The goal of this study is to identify genetic associations for eight quantitative metabolic syndrome (MetS) traits through a meta-analysis across four ethnic groups. Traits were measured in the GENetics of Noninsulin dependent Diabetes Mellitus (GENNID) Study which consists of African-American (families = 73, individuals = 288), European-American (families = 79, individuals = 519), Japanese-American (families = 17, individuals = 132), and Mexican-American (families = 113, individuals = 610) samples. Genome-wide association results from these four ethnic groups were combined using four meta-analysis methods: fixed effects, random effects, TransMeta, and MR-MEGA. We provide an empirical comparison of the four meta-analysis methods from the GENNID results, discuss which types of loci (characterized by allelic heterogeneity) appear to be better detected by each of the four meta-analysis methods in the GENNID Study, and validate our results using previous genetic discoveries. We specifically compare the two transethnic methods, TransMeta and MR-MEGA, and discuss how each transethnic method's framework relates to the types of loci best detected by each method.
Keywords: allelic heterogeneity; genome-wide association; meta-analysis; metabolic syndrome; transethnic.
© 2019 Wiley Periodicals, Inc.
Figures
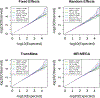
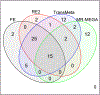




References
-
- Coram MA, Duan Q, Hoffmann TJ, Thornton T, Knowles JW, Johnson NA, … Tang H (2013). Genome-wide Characterization of Shared and Distinct Genetic Components that Influence Blood Lipid Levels in Ethnically Diverse Human Populations. The American Journal of Human Genetics, 92(6), 904–916. 10.1016/j.ajhg.2013.04.025 - DOI - PMC - PubMed

