ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants
- PMID: 31648317
- PMCID: PMC6849945
- DOI: 10.1182/bloodadvances.2019000644
ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants
Abstract
Standardized variant curation is essential for clinical care recommendations for patients with inherited disorders. Clinical Genome Resource (ClinGen) variant curation expert panels are developing disease-associated gene specifications using the 2015 American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) guidelines to reduce curation discrepancies. The ClinGen Myeloid Malignancy Variant Curation Expert Panel (MM-VCEP) was created collaboratively between the American Society of Hematology and ClinGen to perform gene- and disease-specific modifications for inherited myeloid malignancies. The MM-VCEP began optimizing ACMG/AMP rules for RUNX1 because many germline variants have been described in patients with familial platelet disorder with a predisposition to acute myeloid leukemia, characterized by thrombocytopenia, platelet functional/ultrastructural defects, and a predisposition to hematologic malignancies. The 28 ACMG/AMP codes were tailored for RUNX1 variants by modifying gene/disease specifications, incorporating strength adjustments of existing rules, or both. Key specifications included calculation of minor allele frequency thresholds, formulating a semi-quantitative approach to counting multiple independent variant occurrences, identifying functional domains and mutational hotspots, establishing functional assay thresholds, and characterizing phenotype-specific guidelines. Preliminary rules were tested by using a pilot set of 52 variants; among these, 50 were previously classified as benign/likely benign, pathogenic/likely pathogenic, variant of unknown significance (VUS), or conflicting interpretations (CONF) in ClinVar. The application of RUNX1-specific criteria resulted in a reduction in CONF and VUS variants by 33%, emphasizing the benefit of gene-specific criteria and sharing internal laboratory data.
© 2019 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: S.E.P. is a member of the scientific advisory panel of Baylor Genetics Laboratories. L.A.G. is a member of the scientific advisory board for Invitae, Inc., and receives royalties from UpToDate, Inc. L.Z. received honoraria from Future Technology Research, LLC, Roche Diagnostics Asia Pacific, BGI, and Illumina. A family member of L.Z. has a leadership position and ownership interest in the Shanghai Genome Center. The remaining authors declare no competing financial interests.
Figures

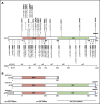


References
-
- Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. - PMC - PubMed
-
- Pepin MG, Murray ML, Bailey S, Leistritz-Kessler D, Schwarze U, Byers PH. The challenge of comprehensive and consistent sequence variant interpretation between clinical laboratories. Genet Med. 2016;18(1):20-24. - PubMed
-
- Amendola LM, Jarvik GP, Leo MC, et al. . Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium [published correction appears in Am J Hum Genet. 2016;99(1):247]. Am J Hum Genet. 2016;98(6):1067-1076. - PMC - PubMed

