Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis
- PMID: 31649238
- PMCID: PMC6813355
- DOI: 10.1038/s41467-019-12412-1
Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis
Abstract
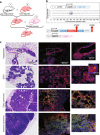
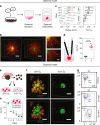
Hypoxia is known to be detrimental in cancer and contributes to its development. In this work, we present an approach to fate-map hypoxic cells in vivo in order to determine their cellular response to physiological O2 gradients as well as to quantify their contribution to metastatic spread. We demonstrate the ability of the system to fate-map hypoxic cells in 2D, and in 3D spheroids and organoids. We identify distinct gene expression patterns in cells that experienced intratumoral hypoxia in vivo compared to cells exposed to hypoxia in vitro. The intratumoral hypoxia gene-signature is a better prognostic indicator for distant metastasis-free survival. Post-hypoxic tumor cells have an ROS-resistant phenotype that provides a survival advantage in the bloodstream and promotes their ability to establish overt metastasis. Post-hypoxic cells retain an increase in the expression of a subset of hypoxia-inducible genes at the metastatic site, suggesting the possibility of a 'hypoxic memory.'
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

