Genetic mapping and evolutionary analysis of human-expanded cognitive networks
- PMID: 31649260
- PMCID: PMC6813316
- DOI: 10.1038/s41467-019-12764-8
Genetic mapping and evolutionary analysis of human-expanded cognitive networks
Abstract
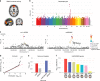
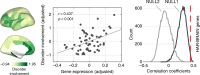
Cognitive brain networks such as the default-mode network (DMN), frontoparietal network, and salience network, are key functional networks of the human brain. Here we show that the rapid evolutionary cortical expansion of cognitive networks in the human brain, and most pronounced the DMN, runs parallel with high expression of human-accelerated genes (HAR genes). Using comparative transcriptomics analysis, we present that HAR genes are differentially more expressed in higher-order cognitive networks in humans compared to chimpanzees and macaques and that genes with high expression in the DMN are involved in synapse and dendrite formation. Moreover, HAR and DMN genes show significant associations with individual variations in DMN functional activity, intelligence, sociability, and mental conditions such as schizophrenia and autism. Our results suggest that the expansion of higher-order functional networks subserving increasing cognitive properties has been an important locus of genetic changes in recent human brain evolution.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Preuss, T. M. Chapter 8: The human brain: evolution and distinctive features. In: On Human Nature.(Academic Press, 2017).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

