Plant-Made Bet v 1 for Molecular Diagnosis
- PMID: 31649716
- PMCID: PMC6795700
- DOI: 10.3389/fpls.2019.01273
Plant-Made Bet v 1 for Molecular Diagnosis
Abstract
Allergic disease diagnosis is currently experiencing a breakthrough due to the use of allergenic molecules in serum-based assays rather than allergen extracts in skin tests. The former methodology is considered a very innovative technology compared with the latter, since it is characterized by flexibility and adaptability to the patient's clinical history and to microtechnology, allowing multiplex analysis. Molecular-based analysis requires pure allergens to detect IgE sensitization, and a major goal, to maintain the diagnosis cost-effective, is to limit their production costs. In addition, for the production of recombinant eukaryotic proteins similar to natural ones, plant-based protein production is preferred to bacterial-based systems due to its ability to perform most of the post-translational modifications of eukaryotic molecules. In this framework, Plant Molecular Farming (PMF) may be useful, being a production platform able to produce complex recombinant proteins in short time-frames at low cost. As a proof of concept, PMF has been exploited for the production of Bet v 1a, a major allergen associated with birch (Betula verrucosa) pollen allergy. Bet v 1a has been produced using two different transient expression systems in Nicotiana benthamiana plants, purified and used in a new generation multiplex allergy diagnosis system, the patient-Friendly Allergen nano-BEad Array (FABER). Plant-made Bet v 1a is immunoreactive, binding IgE and inhibiting IgE-binding to the Escherichia coli expressed allergen currently available in the FABER test, thus suggesting an overall similar though non-overlapping immune activity compared with the E. coli expressed form.
Keywords: IgE; allergen; molecular farming; structure homology; transient expression.
Copyright © 2019 Santoni, Ciardiello, Zampieri, Pezzotti, Giangrieco, Rafaiani, Ciancamerla, Mari and Avesani.
Figures




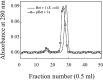
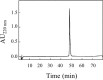





References
-
- Bernardi M. L., Giangrieco I, Camardella L., Ferrara R., Palazzo P., Panico M. R., et al. (2011). Allergenic lipid transfer proteins from plant-derived foods do not immunologically and clinically behave homogeneously: the kiwifruit LTP as a model. PLoS One 6, e27856. 10.1371/journal.pone.0027856. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

