Skeletal muscle mTORC1 regulates neuromuscular junction stability
- PMID: 31651100
- PMCID: PMC7015238
- DOI: 10.1002/jcsm.12496
Skeletal muscle mTORC1 regulates neuromuscular junction stability
Abstract
Background: Skeletal muscle is a plastic tissue that can adapt to different stimuli. It is well established that Mammalian Target of Rapamycin Complex 1 (mTORC1) signalling is a key modulator in mediating increases in skeletal muscle mass and function. However, the role of mTORC1 signalling in adult skeletal muscle homeostasis is still not well defined.
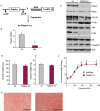
Methods: Inducible, muscle-specific Raptor and mTOR k.o. mice were generated. Muscles at 1 and 7 months after deletion were analysed to assess muscle histology and muscle force.
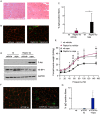
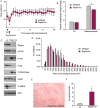
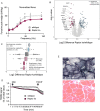
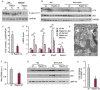
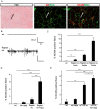
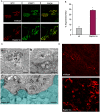
Results: We found no change in muscle size or contractile properties 1 month after deletion. Prolonging deletion of Raptor to 7 months, however, leads to a very marked phenotype characterized by weakness, muscle regeneration, mitochondrial dysfunction, and autophagy impairment. Unexpectedly, reduced mTOR signalling in muscle fibres is accompanied by the appearance of markers of fibre denervation, like the increased expression of the neural cell adhesion molecule (NCAM). Both muscle-specific deletion of mTOR or Raptor, or the use of rapamycin, was sufficient to induce 3-8% of NCAM-positive fibres (P < 0.01), muscle fibrillation, and neuromuscular junction (NMJ) fragmentation in 24% of examined fibres (P < 0.001). Mechanistically, reactivation of autophagy with the small peptide Tat-beclin1 is sufficient to prevent mitochondrial dysfunction and the appearance of NCAM-positive fibres in Raptor k.o. muscles.
Conclusions: Our study shows that mTOR signalling in skeletal muscle fibres is critical for maintaining proper fibre innervation, preserving the NMJ structure in both the muscle fibre and the motor neuron. In addition, considering the beneficial effects of exercise in most pathologies affecting the NMJ, our findings suggest that part of these beneficial effects of exercise are through the well-established activation of mTORC1 in skeletal muscle during and after exercise.
Keywords: Autophagy; Mitochondrial dysfunction; NMJ; mTOR.
© 2019 The Authors. Journal of Cachexia, Sarcopenia and Muscle published by John Wiley & Sons Ltd on behalf of the Society on Sarcopenia, Cachexia and Wasting Disorders.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Marabita M, Baraldo M, Solagna F, Ceelen JJ, Sartori R, Nolte H, et al. S6K1 Is required for increasing skeletal muscle force during hypertrophy. Cell Rep 2016;17:501–513. - PubMed
-
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007;6:458–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

