HLA Heterozygote Advantage against HIV-1 Is Driven by Quantitative and Qualitative Differences in HLA Allele-Specific Peptide Presentation
- PMID: 31651980
- PMCID: PMC7038656
- DOI: 10.1093/molbev/msz249
HLA Heterozygote Advantage against HIV-1 Is Driven by Quantitative and Qualitative Differences in HLA Allele-Specific Peptide Presentation
Abstract
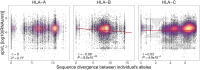
Pathogen-mediated balancing selection is regarded as a key driver of host immunogenetic diversity. A hallmark for balancing selection in humans is the heterozygote advantage at genes of the human leukocyte antigen (HLA), resulting in improved HIV-1 control. However, the actual mechanism of the observed heterozygote advantage is still elusive. HLA heterozygotes may present a broader array of antigenic viral peptides to immune cells, possibly resulting in a more efficient cytotoxic T-cell response. Alternatively, heterozygosity may simply increase the chance to carry the most protective HLA alleles, as individual HLA alleles are known to differ substantially in their association with HIV-1 control. Here, we used data from 6,311 HIV-1-infected individuals to explore the relative contribution of quantitative and qualitative aspects of peptide presentation in HLA heterozygote advantage against HIV. Screening the entire HIV-1 proteome, we observed that heterozygous individuals exhibited a broader array of HIV-1 peptides presented by their HLA class I alleles. In addition, viral load was negatively correlated with the breadth of the HIV-1 peptide repertoire bound by an individual's HLA variants, particularly at HLA-B. This suggests that heterozygote advantage at HLA-B is at least in part mediated by quantitative peptide presentation. We also observed higher HIV-1 sequence diversity among HLA-B heterozygous individuals, suggesting stronger evolutionary pressure from HLA heterozygosity. However, HLA heterozygotes were also more likely to carry certain HLA alleles, including the highly protective HLA-B*57:01 variant, indicating that HLA heterozygote advantage ultimately results from a combination of quantitative and qualitative effects in antigen presentation.
Keywords: MHC evolution; antigen presentation; divergent allele advantage; human leukocyte antigen; major histocompatibility complex; pathogen-mediated balancing selection.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, Ling KL, Barnardo M, Goldthorpe S.. 2003. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet. 12(6):647–656. - PubMed
-
- Apanius V, Penn D, Slev PR, Ruff LR, Potts WK.. 1997. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 17(2):179.. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

