Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance
- PMID: 31653018
- PMCID: PMC6896193
- DOI: 10.3390/cancers11111638
Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance
Abstract
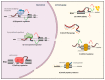
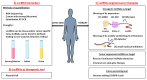
Acute Myeloid Leukemia (AML) is the most common form of leukemia in adults with an incidence of 4.3 per 100,000 cases per year. Historically, the identification of genetic alterations in AML focused on protein-coding genes to provide biomarkers and to understand the molecular complexity of AML. Despite these findings and because of the heterogeneity of this disease, questions as to the molecular mechanisms underlying AML development and progression remained unsolved. Recently, transcriptome-wide profiling approaches have uncovered a large family of long noncoding RNAs (lncRNAs). Larger than 200 nucleotides and with no apparent protein coding potential, lncRNAs could unveil a new set of players in AML development. Originally considered as dark matter, lncRNAs have critical roles to play in the different steps of gene expression and thus affect cellular homeostasis including proliferation, survival, differentiation, migration or genomic stability. Consequently, lncRNAs are found to be differentially expressed in tumors, notably in AML, and linked to the transformation of healthy cells into leukemic cells. In this review, we aim to summarize the knowledge concerning lncRNAs functions and implications in AML, with a particular emphasis on their prognostic and therapeutic potential.
Keywords: acute myeloid leukemia; biomarkers; long noncoding RNA.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures

Similar articles
-
The role of long noncoding RNAs in the diagnosis, prognosis and therapeutic biomarkers of acute myeloid leukemia.Ann Hematol. 2024 Dec;103(12):4931-4942. doi: 10.1007/s00277-024-05987-3. Epub 2024 Sep 12. Ann Hematol. 2024. PMID: 39264436 Review.
-
Genome-wide characterization of lncRNAs in acute myeloid leukemia.Brief Bioinform. 2018 Jul 20;19(4):627-635. doi: 10.1093/bib/bbx007. Brief Bioinform. 2018. PMID: 28203711 Free PMC article.
-
The tip of the iceberg-The roles of long noncoding RNAs in acute myeloid leukemia.Wiley Interdiscip Rev RNA. 2023 Nov-Dec;14(6):e1796. doi: 10.1002/wrna.1796. Epub 2023 Jun 2. Wiley Interdiscip Rev RNA. 2023. PMID: 37267628 Free PMC article. Review.
-
Long noncoding RNAs in acute myeloid leukemia: biomarkers, prognostic indicators, and treatment potential.Cancer Cell Int. 2025 Apr 5;25(1):131. doi: 10.1186/s12935-025-03763-5. Cancer Cell Int. 2025. PMID: 40188050 Free PMC article. Review.
-
Long noncoding RNAs: pivotal regulators in acute myeloid leukemia.Exp Hematol Oncol. 2016 Dec 12;5:30. doi: 10.1186/s40164-016-0059-9. eCollection 2015. Exp Hematol Oncol. 2016. PMID: 27999732 Free PMC article. Review.
Cited by
-
Expression and Alteration Value of Long Noncoding RNA AB073614 and FER1L4 in Patients with Acute Myeloid Leukemia (AML).Asian Pac J Cancer Prev. 2023 Jul 1;24(7):2271-2277. doi: 10.31557/APJCP.2023.24.7.2271. Asian Pac J Cancer Prev. 2023. PMID: 37505756 Free PMC article.
-
Identification of Long Non-Coding RNA SNHG Family as Promising Prognostic Biomarkers in Acute Myeloid Leukemia.Onco Targets Ther. 2020 Aug 24;13:8441-8450. doi: 10.2147/OTT.S265853. eCollection 2020. Onco Targets Ther. 2020. PMID: 32922034 Free PMC article.
-
Long noncoding RNA HOXA-AS2 functions as an oncogene by binding to EZH2 and suppressing LATS2 in acute myeloid leukemia (AML).Cell Death Dis. 2020 Dec 2;11(12):1025. doi: 10.1038/s41419-020-03193-3. Cell Death Dis. 2020. PMID: 33268767 Free PMC article.
-
Computational analysis of heat shock proteins and ferroptosis-associated lncRNAs to predict prognosis in acute myeloid leukemia patients.Front Genet. 2023 Aug 1;14:1218276. doi: 10.3389/fgene.2023.1218276. eCollection 2023. Front Genet. 2023. PMID: 37600655 Free PMC article.
-
The DNA methylation landscape of hematological malignancies: an update.Mol Oncol. 2020 Aug;14(8):1616-1639. doi: 10.1002/1878-0261.12744. Epub 2020 Jul 3. Mol Oncol. 2020. PMID: 32526054 Free PMC article. Review.
References
-
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. Molecular biology: The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

