WRNIP1 Protects Reversed DNA Replication Forks from SLX4-Dependent Nucleolytic Cleavage
- PMID: 31654852
- PMCID: PMC6820244
- DOI: 10.1016/j.isci.2019.10.010
WRNIP1 Protects Reversed DNA Replication Forks from SLX4-Dependent Nucleolytic Cleavage
Abstract
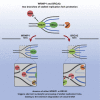
During DNA replication stress, stalled replication forks need to be stabilized to prevent fork collapse and genome instability. The AAA + ATPase WRNIP1 (Werner Helicase Interacting Protein 1) has been implicated in the protection of stalled replication forks from nucleolytic degradation, but the underlying molecular mechanism has remained unclear. Here we show that WRNIP1 exerts its protective function downstream of fork reversal. Unexpectedly though, WRNIP1 is not part of the well-studied BRCA2-dependent branch of fork protection but seems to protect the junction point of reversed replication forks from SLX4-mediated endonucleolytic degradation, possibly by directly binding to reversed replication forks. This function is specific to the shorter, less abundant, and less conserved variant of WRNIP1. Overall, our data suggest that in the absence of BRCA2 and WRNIP1 different DNA substrates are generated at reversed forks but that nascent strand degradation in both cases depends on the activity of exonucleases and structure-specific endonucleases.
Keywords: Biological Sciences; Cell Biology; Molecular Biology.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., De Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014;16:281–291. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

