Feedback to the central dogma: cytoplasmic mRNA decay and transcription are interdependent processes
- PMID: 31656086
- PMCID: PMC6871655
- DOI: 10.1080/10409238.2019.1679083
Feedback to the central dogma: cytoplasmic mRNA decay and transcription are interdependent processes
Abstract
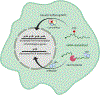
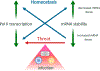
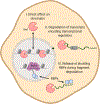
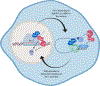
Transcription and RNA decay are key determinants of gene expression; these processes are typically considered as the uncoupled beginning and end of the messenger RNA (mRNA) lifecycle. Here we describe the growing number of studies demonstrating interplay between these spatially disparate processes in eukaryotes. Specifically, cells can maintain mRNA levels by buffering against changes in mRNA stability or transcription, and can also respond to virally induced accelerated decay by reducing RNA polymerase II gene expression. In addition to these global responses, there is also evidence that mRNAs containing a premature stop codon can cause transcriptional upregulation of homologous genes in a targeted fashion. In each of these systems, RNA binding proteins (RBPs), particularly those involved in mRNA degradation, are critical for cytoplasmic to nuclear communication. Although their specific mechanistic contributions are yet to be fully elucidated, differential trafficking of RBPs between subcellular compartments are likely to play a central role in regulating this gene expression feedback pathway.
Keywords: NITC; Xrn1; genetic buffering; host shutoff; mRNA decay; transcription.
Figures