Further evidence of involvement of TMEM132E in autosomal recessive nonsyndromic hearing impairment
- PMID: 31656313
- PMCID: PMC8216908
- DOI: 10.1038/s10038-019-0691-4
Further evidence of involvement of TMEM132E in autosomal recessive nonsyndromic hearing impairment
Erratum in
-
Correction: Further evidence of involvement of TMEM132E in autosomal recessive nonsyndromic hearing impairment.J Hum Genet. 2022 Mar;67(3):181. doi: 10.1038/s10038-021-00951-9. J Hum Genet. 2022. PMID: 34526652 Free PMC article. No abstract available.
Abstract
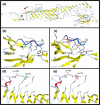
Autosomal-recessive (AR) nonsyndromic hearing impairment (NSHI) displays a high degree of genetic heterogeneity with >100 genes identified. Recently, TMEM132E, which is highly expressed in inner hair cells, was suggested as a novel ARNSHI gene for DFNB99. A missense variant c.1259G>A: p.(Arg420Gln) in TMEM132E was identified that segregated with ARNSHI in a single Chinese family with two affected members. In the present study, a family of Pakistani origin with prelingual profound sensorineural hearing impairment displaying AR mode of inheritance was investigated via exome and Sanger sequencing. Compound heterozygous variants c.382G>T: p.(Ala128Ser) and c.2204C>T: p.(Pro735Leu) in TMEM132E were observed in affected but not in unaffected family members. TMEM132E variants identified in this and the previously reported ARNSHI family are located in the extracellular domain. In conclusion, we present a second ARNSHI family with TMEM132E variants which strengthens the evidence of the involvement of this gene in the etiology of ARNSHI.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Cheng L, Gong Y, Liu Q, Chen B, Guo C, Li J, et al. Gene mapping of a nonsyndromic hearing impairmint family. Chin J Med Genet. 2003;20:89–93. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
