ATP7B Binds Ruthenium(II) p-Cymene Half-Sandwich Complexes: Role of Steric Hindrance and Ru-I Coordination in Rescuing the Sequestration
- PMID: 31657924
- PMCID: PMC7165018
- DOI: 10.1021/acs.inorgchem.9b02780
ATP7B Binds Ruthenium(II) p-Cymene Half-Sandwich Complexes: Role of Steric Hindrance and Ru-I Coordination in Rescuing the Sequestration
Abstract
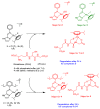
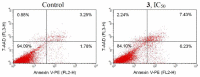
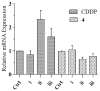
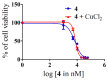
Ruthenium(II/III) complexes are predicted to be efficient alternatives to platinum drug-resistant cancers but have never been investigated for sequestration and efflux by Cu-ATPases (ATP7A or ATP7B) overexpressed in resistant cancer cells, although a major cause of platinum drug resistance is found to be sequestration of platinum chemotherapeutic agents by thiol donors glutathione (GSH) or the Cys-X-X-Cys (CXXC) motifs in the Cu-ATPases in cytosol. Here, we show for the first time that ATP7B efficiently sequesters ruthenium(II) η6-p-cymene complexes. We present seven complexes, [RuII(η6-p-cym)(L)X](PF6) (1-7; L = L1-L3, X = Cl, Br, and I), out of which two resists deactivation by the cellular thiol, glutathione (GSH). The results show that Ru-I coordination and a moderate steric factor increase resistance to GSH and the CXXC motif. RuII-I-coordinated 3 and 7 showed resistance to sequestration by ATP7B. 3 displays highest resistance against GSH and does not trigger ATP7B trafficking in the liver cancer cell line. It escapes ATP7B-mediated sequestration and triggers apoptosis. Thus, with a suitable bidentate ligand and iodido leaving group, RuII(η6-p-cym) complexes may display strong kinetic inertness to inhibit the ATP7B detoxification pathway. Inductively coupled plasma mass spectrometry data show higher retention of 3 and 7 inside the cell with time compared to 4, supporting ATP7B-mediated sequestration.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Noffke AL, Habtemariam A, Pizarro AM, Sadler PJ. Designing organometallic compounds for catalysis and therapy. Chem Commun. 2012;48(43):5219–5246. - PubMed
-
- Hanif M, Hartinger CG. Anticancer metallodrugs: where is the next cisplatin? Future Med Chem. 2018;10(6):615–617. - PubMed
-
- Kenny RG, Marmion CJ. Toward Multi-Targeted Platinum and Ruthenium Drugs-A New Paradigm in Cancer Drug Treatment Regimens? Chem Rev. 2019;119(2):1058–1137. - PubMed
-
- Murray BS, Menin L, Scopelliti R, Dyson PJ. Conformational control of anticancer activity: the application of arene-linked dinuclear ruthenium(ii) organometallics. Chem Sci. 2014;5(6):2536–2545.
-
- Clavel CM, Paunescu E, Nowak-Sliwinska P, Griffioen AW, Scopelliti R, Dyson PJ. Modulating the Anticancer Activity of Ruthenium(II)-Arene Complexes. J Med Chem. 2015;58(8):3356–3365. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

