Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs
- PMID: 31659311
- PMCID: PMC6874898
- DOI: 10.1038/s41557-019-0351-5
Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs
Abstract
A fundamental challenge in chemical biology and medicine is to understand and expand the fraction of the human proteome that can be targeted by small molecules. We recently described a strategy that integrates fragment-based ligand discovery with chemical proteomics to furnish global portraits of reversible small-molecule/protein interactions in human cells. Excavating clear structure-activity relationships from these 'ligandability' maps, however, was confounded by the distinct physicochemical properties and corresponding overall protein-binding potential of individual fragments. Here, we describe a compelling solution to this problem by introducing a next-generation set of fully functionalized fragments differing only in absolute stereochemistry. Using these enantiomeric probe pairs, or 'enantioprobes', we identify numerous stereoselective protein-fragment interactions in cells and show that these interactions occur at functional sites on proteins from diverse classes. Our findings thus indicate that incorporating chirality into fully functionalized fragment libraries provides a robust and streamlined method to discover ligandable proteins in cells.
Conflict of interest statement
Competing Interests
The authors declare competing financial interests. B.F.C. is a founder and advisor to Vividion Therapeutics, a biotechnology company interested in using chemical proteomic methods to develop small-molecule drugs to treat human disease. C.G.P. serves as a consultant for Vividion Therapeutics.
Figures
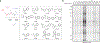


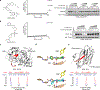

Comment in
-
Ligandable proteome mapping.Nat Methods. 2019 Dec;16(12):1204. doi: 10.1038/s41592-019-0673-7. Nat Methods. 2019. PMID: 31780831 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

