The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy
- PMID: 31659324
- PMCID: PMC6858879
- DOI: 10.1038/s41588-019-0517-5
The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy
Abstract
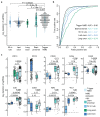
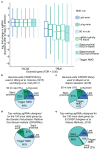
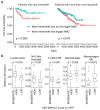
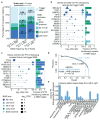
Premature termination codons (PTCs) can result in the production of truncated proteins or the degradation of messenger RNAs by nonsense-mediated mRNA decay (NMD). Which of these outcomes occurs can alter the effect of a mutation, with the engagement of NMD being dependent on a series of rules. Here, by applying these rules genome-wide to obtain a resource called NMDetective, we explore the impact of NMD on genetic disease and approaches to therapy. First, human genetic diseases differ in whether NMD typically aggravates or alleviates the effects of PTCs. Second, failure to trigger NMD is a cause of ineffective gene inactivation by CRISPR-Cas9 gene editing. Finally, NMD is a determinant of the efficacy of cancer immunotherapy, with only frameshifted transcripts that escape NMD predicting a response. These results demonstrate the importance of incorporating the rules of NMD into clinical decision-making. Moreover, they suggest that inhibiting NMD may be effective in enhancing cancer immunotherapy.
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–77. - PubMed
-
- Silva AL, Romao L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499–505. - PubMed
-
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

