Domainoid: domain-oriented orthology inference
- PMID: 31660857
- PMCID: PMC6816169
- DOI: 10.1186/s12859-019-3137-2
Domainoid: domain-oriented orthology inference
Abstract
Background: Orthology inference is normally based on full-length protein sequences. However, most proteins contain independently folding and recurring regions, domains. The domain architecture of a protein is vital for its function, and recombination events mean individual domains can have different evolutionary histories. It has previously been shown that orthologous proteins may differ in domain architecture, creating challenges for orthology inference methods operating on full-length sequences. We have developed Domainoid, a new tool aiming to overcome these challenges faced by full-length orthology methods by inferring orthology on the domain level. It employs the InParanoid algorithm on single domains separately, to infer groups of orthologous domains.
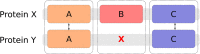
Results: This domain-oriented approach allows detection of discordant domain orthologs, cases where different domains on the same protein have different evolutionary histories. In addition to domain level analysis, protein level orthology based on the fraction of domains that are orthologous can be inferred. Domainoid orthology assignments were compared to those yielded by the conventional full-length approach InParanoid, and were validated in a standard benchmark.
Conclusions: Our results show that domain-based orthology inference can reveal many orthologous relationships that are not found by full-length sequence approaches.
Availability: https://bitbucket.org/sonnhammergroup/domainoid/.
Keywords: Domain ortholog; Orthology; Protein domain.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

