Proper secretion of the serpin antithrombin relies strictly on thiol-dependent quality control
- PMID: 31662433
- PMCID: PMC6916506
- DOI: 10.1074/jbc.RA119.010450
Proper secretion of the serpin antithrombin relies strictly on thiol-dependent quality control
Abstract
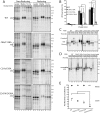
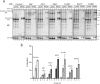
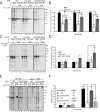
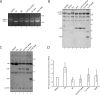
The protein quality control machinery of the endoplasmic reticulum (ERQC) ensures that client proteins are properly folded. ERQC substrates may be recognized as nonnative by the presence of exposed hydrophobic surfaces, free thiols, or processed N-glycans. How these features dictate which ERQC pathways engage a given substrate is poorly understood. Here, using metabolic labeling, immunoprecipitations, various biochemical assays, and the human serpin antithrombin III (ATIII) as a model, we explored the role of ERQC systems in mammalian cells. Although ATIII has N-glycans and a hydrophobic core, we found that its quality control depended solely on free thiol content. Mutagenesis of all six Cys residues in ATIII to Ala resulted in its efficient secretion even though the product was not natively folded. ATIII variants with free thiols were retained in the endoplasmic reticulum but not degraded. These results provide insight into the hierarchy of ERQC systems and reveal a fundamental vulnerability of ERQC in a case of reliance on the thiol-dependent quality control pathway.
Keywords: ER-mediated protein quality control (ERQC); N-linked glycosylation; UPR; disulfide; endoplasmic reticulum (ER); endoplasmic reticulum-associated protein degradation (ERAD); protein folding; protein homeostasis; protein maturation; serpin; unfolded protein response (UPR).
© 2019 Adams et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

