Maintenance of Sympatric and Allopatric Populations in Free-Living Terrestrial Bacteria
- PMID: 31662456
- PMCID: PMC6819660
- DOI: 10.1128/mBio.02361-19
Maintenance of Sympatric and Allopatric Populations in Free-Living Terrestrial Bacteria
Abstract
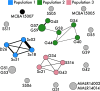
For free-living bacteria and archaea, the equivalent of the biological species concept does not exist, creating several obstacles to the study of the processes contributing to microbial diversification. These obstacles are particularly high in soil, where high bacterial diversity inhibits the study of closely related genotypes and therefore the factors structuring microbial populations. Here, we isolated strains within a single Curtobacterium ecotype from surface soil (leaf litter) across a regional climate gradient and investigated the phylogenetic structure, recombination, and flexible gene content of this genomic diversity to infer patterns of gene flow. Our results indicate that microbial populations are delineated by gene flow discontinuities, with distinct populations cooccurring at multiple sites. Bacterial population structure was further delineated by genomic features allowing for the identification of candidate genes possibly contributing to local adaptation. These results suggest that the genetic structure within this bacterium is maintained both by ecological specialization in localized microenvironments (isolation by environment) and by dispersal limitation between geographic locations (isolation by distance).IMPORTANCE Due to the promiscuous exchange of genetic material and asexual reproduction, delineating microbial species (and, by extension, populations) remains challenging. Because of this, the vast majority of microbial studies assessing population structure often compare divergent strains from disparate environments under varied selective pressures. Here, we investigated the population structure within a single bacterial ecotype, a unit equivalent to a eukaryotic species, defined as highly clustered genotypic and phenotypic strains with the same ecological niche. Using a combination of genomic and computational analyses, we assessed the phylogenetic structure, extent of recombination, and flexible gene content of this genomic diversity to infer patterns of gene flow. To our knowledge, this study is the first to do so for a dominant soil bacterium. Our results indicate that bacterial soil populations, similarly to those in other environments, are structured by gene flow discontinuities and exhibit distributional patterns consistent with both isolation by distance and isolation by environment. Thus, both dispersal limitation and local environments contribute to the divergence among closely related soil bacteria as observed in macroorganisms.
Keywords: Curtobacterium; ecotype; gene flow; microbial ecology; population structure.
Figures



References
-
- Mayr E. 2001. What evolution is. Science Masters Series. Basic Books, New York, NY.
-
- Chase AB, Martiny J. 2018. The importance of resolving biogeographic patterns of microbial microdiversity. Microbiol Aust 39:5–8.
