Top-Down ETD-MS Provides Unreliable Quantitation of Methionine Oxidation
- PMID: 31662705
- PMCID: PMC6808186
- DOI: 10.7171/jbt.19-3004-002
Top-Down ETD-MS Provides Unreliable Quantitation of Methionine Oxidation
Abstract
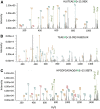
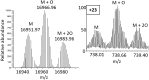
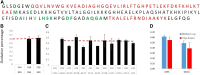
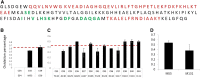
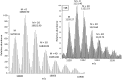
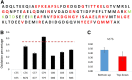
Methionine oxidation plays a critical role in many processes of biologic and biomedical importance, including cellular redox responses and stability of protein pharmaceuticals. Bottom-up methods for analysis of methionine oxidation can suffer from incomplete sequence coverage, as well as an inability to readily detect correlated oxidation between 2 or more methionines. However, the methodology for quantifying protein oxidation in top-down analyses is lacking. Previous work has shown that electron transfer dissociation (ETD)-based tandem mass spectrometry (MS/MS) fragmentation offers accurate and precise quantification of amino acid oxidation in peptides, even in complex samples. However, the ability of ETD-based MS/MS fragmentation to accurately quantify amino acid oxidation of proteins in a top-down manner has not been reported. Using apomyoglobin and calmodulin as model proteins, we partially converted methionines into methionine sulfoxide by incubation in H2O2. Using top-down ETD-based fragmentation, we quantified the amount of oxidation of various ETD product ions and compared the quantified values with those from traditional bottom-up analysis. We find that overall quantification of methionine oxidation by top-down MS/MS ranges from good agreement with traditional bottom-up methods to vast differences between the 2 techniques, including missing oxidized product ions and large differences in measured oxidation quantities. Care must be taken in transitioning ETD-based quantitation of oxidation from the peptide level to the intact protein level.
Keywords: hydroxyl radical protein footprinting; post-translational modification; proteomics.
© Association of Biomolecular Resource Facilities.
Figures

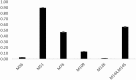
References
-
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
