Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3
- PMID: 31664039
- PMCID: PMC6820781
- DOI: 10.1038/s41467-019-12763-9
Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3
Abstract
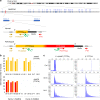
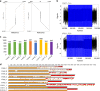
Familial Adult Myoclonic Epilepsy (FAME) is a genetically heterogeneous disorder characterized by cortical tremor and seizures. Intronic TTTTA/TTTCA repeat expansions in SAMD12 (FAME1) are the main cause of FAME in Asia. Using genome sequencing and repeat-primed PCR, we identify another site of this repeat expansion, in MARCH6 (FAME3) in four European families. Analysis of single DNA molecules with nanopore sequencing and molecular combing show that expansions range from 3.3 to 14 kb on average. However, we observe considerable variability in expansion length and structure, supporting the existence of multiple expansion configurations in blood cells and fibroblasts of the same individual. Moreover, the largest expansions are associated with micro-rearrangements occurring near the expansion in 20% of cells. This study provides further evidence that FAME is caused by intronic TTTTA/TTTCA expansions in distinct genes and reveals that expansions exhibit an unexpectedly high somatic instability that can ultimately result in genomic rearrangements.
Conflict of interest statement
Y.D. and M.D.M.A. were employed by Genomic Vision, a company providing diagnostic kits for the detection of genomic rearrangements, at the time of the study. All other authors declare no competing interests.
Figures




References
-
- Striano P, Zara F. Autosomal dominant cortical tremor, myoclonus and epilepsy. Epileptic Disord. 2016;18:139–144. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

