Network vulnerability-based and knowledge-guided identification of microRNA biomarkers indicating platinum resistance in high-grade serous ovarian cancer
- PMID: 31664600
- PMCID: PMC6820656
- DOI: 10.1186/s40169-019-0245-6
Network vulnerability-based and knowledge-guided identification of microRNA biomarkers indicating platinum resistance in high-grade serous ovarian cancer
Abstract
Background: High-grade serous ovarian cancer (HGSC), the most common ovarian carcinoma type, is associated with the highest mortality rate among all gynecological malignancies. As chemoresistance has been demonstrated as the major challenge in improving the prognosis of HGSC patients, we here aimed to identify microRNA (miRNA) biomarkers for predicting platinum resistance and further explore their functions in HGSC.
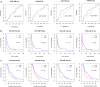
Results: We developed and applied our network vulnerability-based and knowledge-guided bioinformatics model first time for the study of drug-resistance in cancer. Four miRNA biomarkers (miR-454-3p, miR-98-5p, miR-183-5p and miR-22-3p) were identified with potential in stratifying platinum-sensitive and platinum-resistant HGSC patients and predicting prognostic outcome. Among them, miR-454-3p and miR-183-5p were newly discovered to be closely implicated in platinum resistance in HGSC. Functional analyses highlighted crucial roles of the four miRNA biomarkers in platinum resistance through mediating transcriptional regulation, cell proliferation and apoptosis. Moreover, expression patterns of the miRNA biomarkers were validated in both platinum-sensitive and platinum-resistant ovarian cancer cells.
Conclusions: With bioinformatics modeling and analysis, we identified and confirmed four novel putative miRNA biomarkers, miR-454-3p, miR-98-5p, miR-183-5p and miR-22-3p that could serve as indicators of resistance to platinum-based chemotherapy, thereby contributing to the improvement of chemotherapeutic efficiency and optimization of personalized treatments in HGSC.
Keywords: Bioinformatics model; Ovarian cancer; Platinum resistance; miRNA biomarker; miRNA-mRNA regulatory network.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




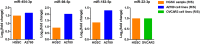
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
