Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology
- PMID: 31664817
- PMCID: PMC7382785
- DOI: 10.1021/acsnano.9b06522
Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology
Abstract
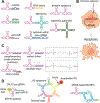
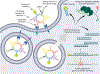
Nucleic acids play a central role in all domains of life, either as genetic blueprints or as regulators of various biochemical pathways. The chemical makeup of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA), generally represented by a sequence of four monomers, also provides precise instructions for folding and higher-order assembly of these biopolymers that, in turn, dictate biological functions. The sequence-based specific 3D structures of nucleic acids led to the development of the directed evolution of oligonucleotides, SELEX (systematic evolution of ligands by exponential enrichment), against a chosen target molecule. Among the variety of functions, selected oligonucleotides named aptamers also allow targeting of cell-specific receptors with antibody-like precision and can deliver functional RNAs without a transfection agent. The advancements in the field of customizable nucleic acid nanoparticles (NANPs) opened avenues for the design of nanoassemblies utilizing aptamers for triggering or blocking cell signaling pathways or using aptamer-receptor combinations to activate therapeutic functionalities. A recent selection of fluorescent aptamers enables real-time tracking of NANP formation and interactions. The aptamers are anticipated to contribute to the future development of technologies, enabling an efficient assembly of functional NANPs in mammalian cells or in vivo. These research topics are of top importance for the field of therapeutic nucleic acid nanotechnology with the promises to scale up mass production of NANPs suitable for biomedical applications, to control the intracellular organization of biological materials to enhance the efficiency of biochemical pathways, and to enhance the therapeutic potential of NANP-based therapeutics while minimizing undesired side effects and toxicities.
Keywords: NANPs; RNA nanotechnology; SELEX; aptamers; exosomes; immunotherapy; nucleic acid delivery; therapeutic nucleic acids.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- Cech TR; Steitz JA The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

