Reconstruction of Dispersal Patterns of Hypervirulent Meningococcal Strains of Serogroup C:cc11 by Phylogenomic Time Trees
- PMID: 31666361
- PMCID: PMC6935922
- DOI: 10.1128/JCM.01351-19
Reconstruction of Dispersal Patterns of Hypervirulent Meningococcal Strains of Serogroup C:cc11 by Phylogenomic Time Trees
Abstract
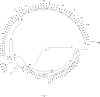
Neisseria meningitidis is one of the few commensal bacteria that can even cause large epidemics of invasive meningococcal disease (IMD). N. meningitis serogroup C belonging to the hypervirulent clonal complex 11 (cc11) represents an important public health threat worldwide. We reconstructed the dispersal patterns of hypervirulent meningococcal strains of serogroup C:cc11 by phylogenomic time trees. In particular, we focused the attention on the epidemic dynamics of C:P1.5.1,10-8:F3-6;ST-11(cc11) meningococci causing outbreaks, as occurred in the Tuscany region, Italy, in 2015 to 2016. A phylogeographic analysis was performed through a Bayesian method on 103 Italian and 208 foreign meningococcal genomes. The C:P1.5.1,10-8:F3-6;ST-11(cc11) genotype dated back to 1995 (1992 to 1998) in the United Kingdom. Two main clades of the hypervirulent genotype were identified in Italy. The Tuscany outbreak isolates were included in different clusters in a specific subclade which originated in the United Kingdom around 2011 and was introduced in Tuscany in 2013 to 2014. In this work, phylogeographic analysis allowed the identification of multiple introductions of these strains in several European countries and connections with extra-European areas. Whole-genome sequencing (WGS) combined with phylogeography enables us to track the dissemination of meningococci and their transmission. The C:P1.5.1,10-8:F3-6;ST-11(cc11) genotype analysis revealed how a hypervirulent strain may be introduced in previously naïve areas, causing a large and long-lasting outbreak.
Keywords: Neisseria meningitidis; invasive meningococcal disease; phylogeography; public health surveillance.
Copyright © 2019 Lo Presti et al.
Figures




References
-
- Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA, Corton C, Care R, Poolman JT, Zollinger WD, Frasch CE, Stephens DS, Feavers I, Frosch M, Parkhill J, Vogel U, Quail MA, Bentley SD, Maiden MC. 2013. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis 19:566–573. doi:10.3201/eid1904.111799. - DOI - PMC - PubMed
-
- Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi:10.1073/pnas.95.6.3140. - DOI - PMC - PubMed
-
- Yazdankhah SP, Kriz P, Tzanakaki G, Kremastinou J, Kalmusova J, Musilek M, Alvestad T, Jolley KA, Wilson DJ, McCarthy ND, Caugant DA, Maiden MC. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J Clin Microbiol 42:5146–5153. doi:10.1128/JCM.42.11.5146-5153.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

