Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples
- PMID: 31666364
- PMCID: PMC6935926
- DOI: 10.1128/JCM.00963-19
Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples
Abstract
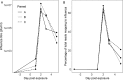
Influenza is a major global public health threat as a result of its highly pathogenic variants, large zoonotic reservoir, and pandemic potential. Metagenomic viral sequencing offers the potential for a diagnostic test for influenza virus which also provides insights on transmission, evolution, and drug resistance and simultaneously detects other viruses. We therefore set out to apply the Oxford Nanopore Technologies sequencing method to metagenomic sequencing of respiratory samples. We generated influenza virus reads down to a limit of detection of 102 to 103 genome copies/ml in pooled samples, observing a strong relationship between the viral titer and the proportion of influenza virus reads (P = 4.7 × 10-5). Applying our methods to clinical throat swabs, we generated influenza virus reads for 27/27 samples with mid-to-high viral titers (cycle threshold [CT ] values, <30) and 6/13 samples with low viral titers (CT values, 30 to 40). No false-positive reads were generated from 10 influenza virus-negative samples. Thus, Nanopore sequencing operated with 83% sensitivity (95% confidence interval [CI], 67 to 93%) and 100% specificity (95% CI, 69 to 100%) compared to the current diagnostic standard. Coverage of full-length virus was dependent on sample composition, being negatively influenced by increased host and bacterial reads. However, at high influenza virus titers, we were able to reconstruct >99% complete sequences for all eight gene segments. We also detected a human coronavirus coinfection in one clinical sample. While further optimization is required to improve sensitivity, this approach shows promise for the Nanopore platform to be used in the diagnosis and genetic analysis of influenza virus and other respiratory viruses.
Keywords: DNA sequencing; Nanopore; diagnosis; diagnostics; epidemiology; influenza; metagenomic; metagenomics; molecular epidemiology; sequencing.
Copyright © 2019 Lewandowski et al.
Figures
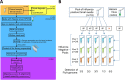
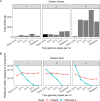
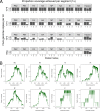

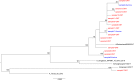
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

