Adipocyte browning and resistance to obesity in mice is induced by expression of ATF3
- PMID: 31667363
- PMCID: PMC6813364
- DOI: 10.1038/s42003-019-0624-y
Adipocyte browning and resistance to obesity in mice is induced by expression of ATF3
Abstract
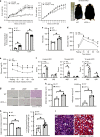
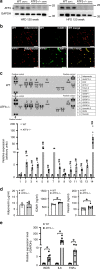
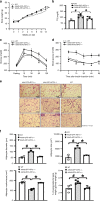
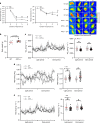
Billions of people have obesity-related metabolic syndromes such as diabetes and hyperlipidemia. Promoting the browning of white adipose tissue has been suggested as a potential strategy, but a drug still needs to be identified. Here, genetic deletion of activating transcription factor 3 (ATF3-/- ) in mice under a high-fat diet (HFD) resulted in obesity and insulin resistance, which was abrogated by virus-mediated ATF3 restoration. ST32da, a synthetic ATF3 inducer isolated from Salvia miltiorrhiza, promoted ATF3 expression to downregulate adipokine genes and induce adipocyte browning by suppressing the carbohydrate-responsive element-binding protein-stearoyl-CoA desaturase-1 axis. Furthermore, ST32da increased white adipose tissue browning and reduced lipogenesis in HFD-induced obese mice. The anti-obesity efficacy of oral ST32da administration was similar to that of the clinical drug orlistat. Our study identified the ATF3 inducer ST32da as a promising therapeutic drug for treating diet-induced obesity and related metabolic disorders.
Keywords: Diabetes; Drug discovery; Medical research; Obesity; Transcription.
© The Author(s) 2019.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

