Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb
- PMID: 31668641
- PMCID: PMC6948188
- DOI: 10.1016/j.immuni.2019.10.001
Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb
Abstract
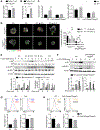
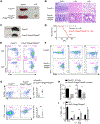
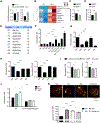
Regulatory T (Treg) cells are critical mediators of immune tolerance whose activity depends upon T cell receptor (TCR) and mTORC1 kinase signaling, but the mechanisms that dictate functional activation of these pathways are incompletely understood. Here, we showed that amino acids license Treg cell function by priming and sustaining TCR-induced mTORC1 activity. mTORC1 activation was induced by amino acids, especially arginine and leucine, accompanied by the dynamic lysosomal localization of the mTOR and Tsc complexes. Rag and Rheb GTPases were central regulators of amino acid-dependent mTORC1 activation in effector Treg (eTreg) cells. Mice bearing RagA-RagB- or Rheb1-Rheb2-deficient Treg cells developed a fatal autoimmune disease and had reduced eTreg cell accumulation and function. RagA-RagB regulated mitochondrial and lysosomal fitness, while Rheb1-Rheb2 enforced eTreg cell suppressive gene signature. Together, these findings reveal a crucial requirement of amino acid signaling for licensing and sustaining mTORC1 activation and functional programming of Treg cells.
Keywords: RagA; RagB; Rheb; Treg cells; amino acids; autoimmunity; eTreg cells; mTOR; metabolism.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
K.-L.G. is a co-founder and has an equity interest in Vivace Therapeutics, Inc.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

